5UFV
 
 | |
4EIR
 
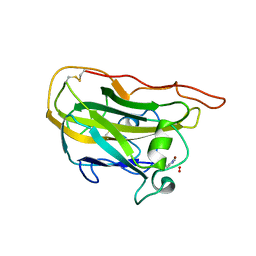 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
6PAS
 
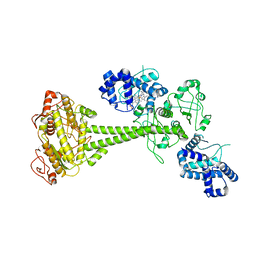 | | Inactive State of Manduca sexta soluble guanylate cyclase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble guanylyl cyclase alpha-1 subunit, Soluble guanylyl cyclase beta-1 subunit | | Authors: | Yokom, A.L, Horst, B.G, Marletta, M.A, Hurley, J.H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife, 8, 2019
|
|
6PAT
 
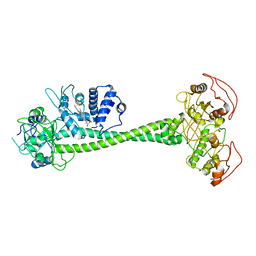 | | Active State of Manduca sexta soluble Guanylate Cyclase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble guanylyl cyclase alpha-1 subunit, Soluble guanylyl cyclase beta-1 subunit | | Authors: | Yokom, A.L, Horst, B.G, Marletta, M.A, Hurley, J.H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife, 8, 2019
|
|
3EEE
 
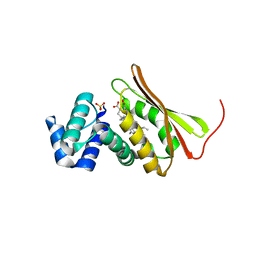 | | Probing the function of heme distortion in the H-NOX family | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Olea Jr, C, Boon, E.M, Pellicena, P, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Probing the function of heme distortion in the H-NOX family.
Acs Chem.Biol., 3, 2008
|
|
3ET6
 
 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | Descriptor: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | Authors: | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
6BDE
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein mutant A71G from K. algicida | | Descriptor: | 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J.J, Hespen, C.W, Marletta, M.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins.
ACS Chem. Biol., 13, 2018
|
|
6BDD
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein from K. algicida | | Descriptor: | 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J.J, Hespen, C.W, Marletta, M.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins.
ACS Chem. Biol., 13, 2018
|
|
5JRU
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein from C. subterraneus | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
5L2V
 
 | | Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, COPPER (II) ION, Chitin-binding protein | | Authors: | Light, S.H, Agostoni, M, Marletta, M.A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes
To Be Published
|
|
4U9G
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9K
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II)NO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9J
 
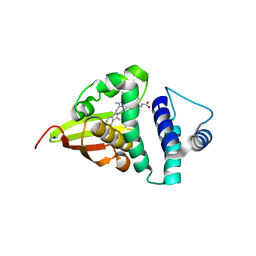 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9B
 
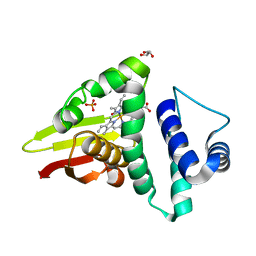 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IT2
 
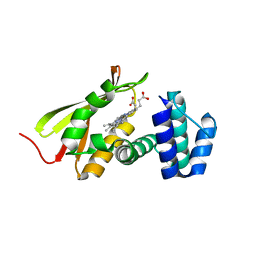 | | Mn(III)-PPIX bound Tt H-NOX | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, Methyl-accepting chemotaxis protein | | Authors: | Winter, M.B, Klemm, P.J, Phillips-Piro, C.M, Raymond, K.M, Marletta, M.A. | | Deposit date: | 2013-01-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Porphyrin-Substituted H-NOX Proteins as High-Relaxivity MRI Contrast Agents.
Inorg.Chem., 52, 2013
|
|
3LAI
 
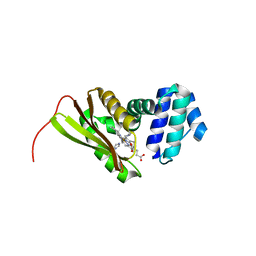 | | Structural insights into the molecular mechanism of H-NOX activation | | Descriptor: | IMIDAZOLE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Olea Jr, C, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural insights into the molecular mechanism of H-NOX activation.
Protein Sci., 19, 2010
|
|
5JRV
 
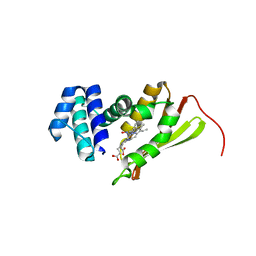 | | Crystal structure of Fe(II) NO-bound H-NOX protein from C. subterraneus | | Descriptor: | IODIDE ION, Methyl-accepting chemotaxis protein, NITRIC OXIDE, ... | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
5JRX
 
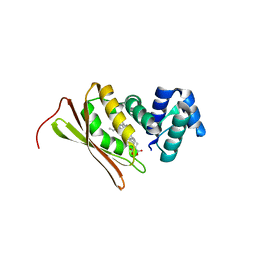 | | Crystal structure of Fe(II) CO-bound H-NOX protein from C. subterraneus | | Descriptor: | CARBON MONOXIDE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
1U4H
 
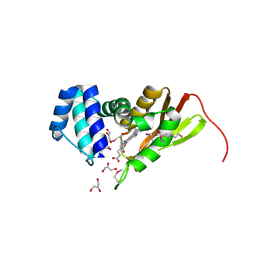 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (oxygen complex) | | Descriptor: | GLYCEROL, Heme-based Methyl-accepting Chemotaxis Protein, OXYGEN MOLECULE, ... | | Authors: | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4FDK
 
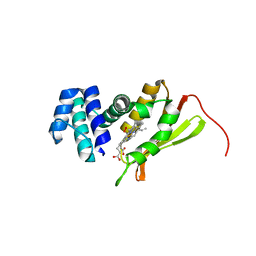 | | F78L Tt H-NOX | | Descriptor: | Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weinert, E.E, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2012-05-28 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin pi-stacking in a heme protein scaffold tunes gas ligand affinity.
J.Inorg.Biochem., 127C, 2013
|
|
3GOF
 
 | | Calmodulin bound to peptide from macrophage nitric oxide synthase | | Descriptor: | CALCIUM ION, Calmodulin, Nitric oxide synthase, ... | | Authors: | Ng, H.L, Greenstein, A, Marletta, M, Wand, A.J, Alber, T. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural diversity in calmodulin recognition of nitric oxide synthases
To be Published
|
|
1U56
 
 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (Water-ligated, ferric form) | | Descriptor: | CHLORIDE ION, Heme-based Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1U55
 
 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (oxygen complex) | | Descriptor: | CHLORIDE ION, Heme-based Methyl-accepting Chemotaxis Protein, OXYGEN MOLECULE, ... | | Authors: | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4EIS
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, PEROXIDE ION, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
