3KPP
 
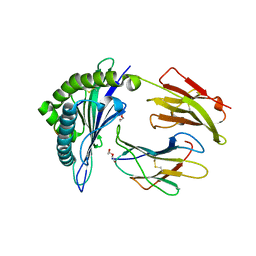 | | Crystal Structure of HLA B*4405 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, EEYLQAFTY, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
1KAC
 
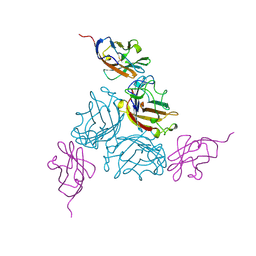 | | KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 IN COMPLEX WITH DOMAIN 1 OF ITS CELLULAR RECEPTOR CAR | | Descriptor: | PROTEIN (COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR), PROTEIN (FIBER KNOB PROTEIN) | | Authors: | Bewley, M.C, Springer, K, Zhang, Y.B, Freimuth, P, Flanagan, J.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR.
Science, 286, 1999
|
|
1K4I
 
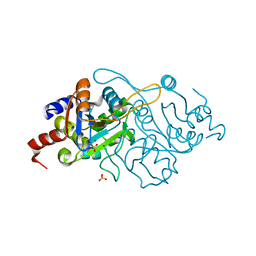 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with two Magnesium ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, MAGNESIUM ION, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
1K5R
 
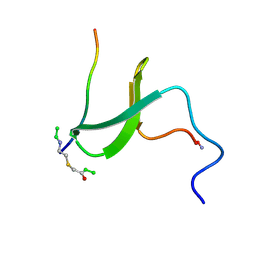 | | YAP65 WW domain S24-Amino-Ethylsulfanyl-Acetic Acid mutant | | Descriptor: | 65 KDA YES-ASSOCIATED PROTEIN, Fragment of WBP-1 | | Authors: | Ferguson, N, Pires, J.R, Toepert, F, Johnson, C.M, Pan, Y.P, Volkmer-Engert, R, Schneider-Mergener, J, Daggett, V, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2001-10-12 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Using flexible loop mimetics to extend phi-value analysis to secondary structure interactions.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K4L
 
 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with two Manganese ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, MANGANESE (II) ION, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
4QB0
 
 | |
4QMD
 
 | | Crystal structure of human envoplakin plakin repeat domain | | Descriptor: | Envoplakin | | Authors: | Mohammed, F, Al-Jassar, C, White, S.A, Fogl, C, Jeeves, M, Knowles, T.J, Odinstova, E, Rodriguez-Zamora, P, Overduin, M, Chidgey, M. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin.
Nat Commun, 7, 2016
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4QNM
 
 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
3L0T
 
 | | Crystal structure of catalytic domain of TACE with hydantoin inhibitor | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and SAR of hydantoin TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1KB7
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
4N0V
 
 | | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332 | | Descriptor: | Glutathione S-transferase, N-terminal domain | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332
TO BE PUBLISHED
|
|
3L35
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
4QG9
 
 | | crystal structure of PKM2-R399E mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
3L3X
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
4QHH
 
 | | Crystal structure of Methanocaldococcus jannaschii tetrameric selecase | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3KPM
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
4N4Z
 
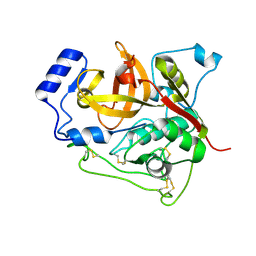 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
4N9V
 
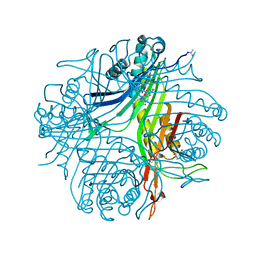 | | High resolution x-ray structure of urate oxidase in complex with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
3L6H
 
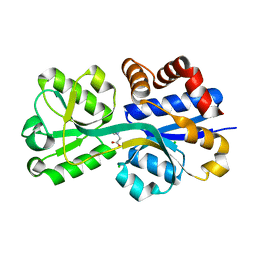 | | Crystal structure of lactococcal OpuAC in its closed-liganded conformation complexed with glycine betaine | | Descriptor: | Betaine ABC transporter permease and substrate binding protein, CHLORIDE ION, TRIMETHYL GLYCINE | | Authors: | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
4MU1
 
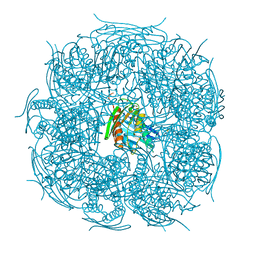 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+, imidazole, and sulfate at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
3LE9
 
 | |
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QL1
 
 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
