3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1KMF
 
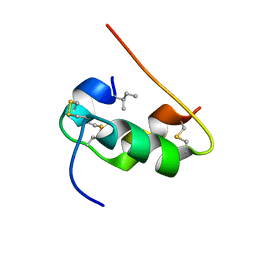 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1J73
 
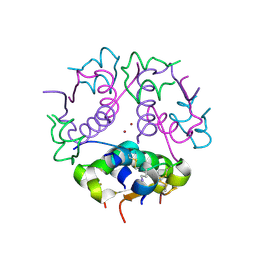 | | Crystal structure of an unstable insulin analog with native activity. | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Wan, Z, Zhao, M, Nakagawa, S, Jia, W, Weiss, M.A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1RWE
 
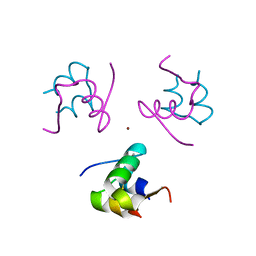 | | Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z, Xu, B, Chu, Y.C, Li, B, Nakagawa, S.H, Qu, Y, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues.
Biochemistry, 43, 2004
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
2HH4
 
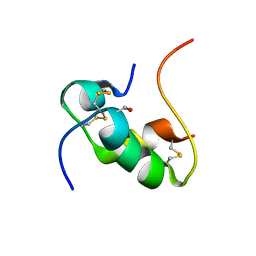 | | NMR structure of human insulin mutant GLY-B8-D-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | insulin A chain, insulin B chain | | Authors: | Hua, Q.X, Nakagawa, S, Hu, S.Q, Jia, W, Weiss, M.A. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Active Conformation of Insulin: Stereospecific modulation of a structural switch in the B chain.
J.Biol.Chem., 281, 2006
|
|
2HHO
 
 | | NMR structure of human insulin mutant GLY-B8-SER, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Nakagawa, S, Hu, S.Q, Jia, W, Weiss, M.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Active Conformation of Insulin: Stereospecific modulation of a structural switch in the B chain.
J.Biol.Chem., 281, 2006
|
|
2JUU
 
 | | allo-ThrA3 DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
2JUM
 
 | | ThrA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-08-31 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
2JUV
 
 | | AbaA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
1K3M
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Xu, B, Hua, Q.-X, Nakagawa, S.H, Jia, W, Chu, Y.-C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-17 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix.
Protein Sci., 11, 2002
|
|
1IPJ
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS COMPLEXES WITH N-ACETYL-D-GLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1IPK
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1T1K
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T1Q
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ABA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin, insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T1P
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-THR, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin, insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
2ZW3
 
 | | Structure of the connexin-26 gap junction channel at 3.5 angstrom resolution | | Descriptor: | Gap junction beta-2 protein | | Authors: | Maeda, S, Nakagawa, S, Suga, M, Yamashita, E, Oshima, A, Fujiyoshi, Y, Tsukihara, T. | | Deposit date: | 2008-12-01 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the connexin 26 gap junction channel at 3.5 A resolution
Nature, 458, 2009
|
|
