8FWI
 
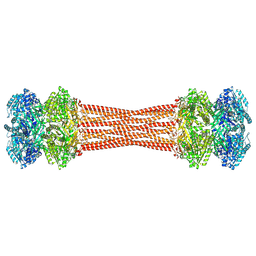 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
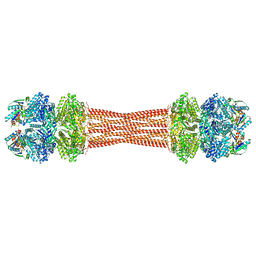 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
1JB0
 
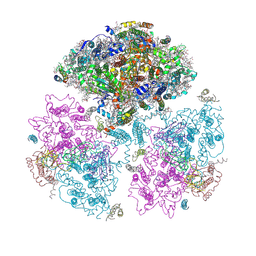 | | Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Jordan, P, Fromme, P, Witt, H.T, Klukas, O, Saenger, W, Krauss, N. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of Cyanobacterial Photosystem I at 2.5 A Resolution
NATURE, 411, 2001
|
|
5FXH
 
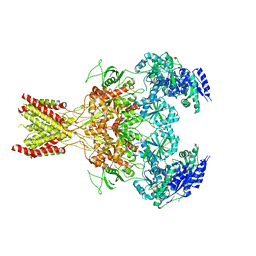 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
7WMC
 
 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
2WC1
 
 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
5AEP
 
 | | Novel pyrrole carboxamide inhibitors of JAK2 as potential treatment of myeloproliferative disorders | | Descriptor: | 1-(5-chloro-2-methylphenyl)-4-(pyrrolo[2,1-f][1,2,4]triazin-4-yl)-1H-pyrrole-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Bertrand, J, Brasca, M.G, Nesi, M, Amboldi, N, Avanzi, N, Bindi, S, Casero, D, Ciomei, M, Colombo, N, Cribioli, S, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Motto, I, Panzeri, A, Gnocchi, P, Donati, D. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Pyrrole Carboxamide Inhibitors of Jak2 as Potential Treatment of Myeloproliferative Disorders.
Bioorg.Med.Chem., 23, 2015
|
|
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
5FXG
 
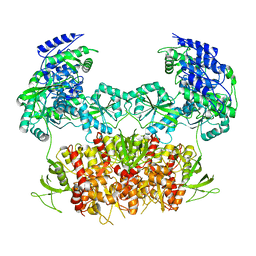 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
1KTP
 
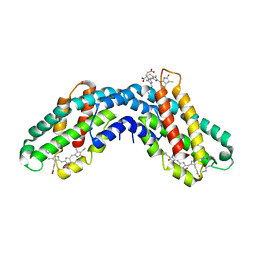 | | Crystal structure of c-phycocyanin of synechococcus vulcanus at 1.6 angstroms | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, ... | | Authors: | Adir, N, Dobrovetsky, E, Lerner, N. | | Deposit date: | 2002-01-17 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structure of c-phycocyanin from the cyanobacterium Synechococcus vulcanus at 1.6 A: insights into the role of solvent molecules in thermal stability and co-factor structure
Biochim.Biophys.Acta, 1556, 2002
|
|
6WZN
 
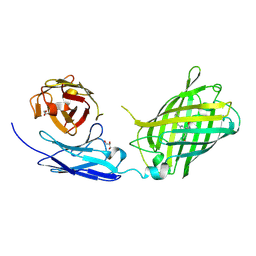 | | Crystal Structure of a Fluorescent Single Chain Fv Chimera | | Descriptor: | Fluorescent Single Chain Fv Chimera, GLYCEROL | | Authors: | Close, D, Velappan, N, Hung, L.W, Naranjo, L, Hemez, C, DeVore, N, McCullough, D, Lillo, A.M, Waldo, G, Bradbury, A.R.M. | | Deposit date: | 2020-05-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Construction, characterization and crystal structure of a fluorescent single-chain Fv chimera.
Protein Eng.Des.Sel., 34, 2021
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
6QMR
 
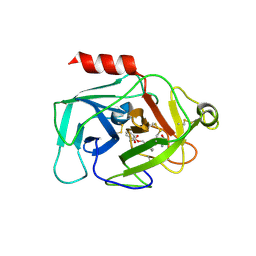 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
6QMT
 
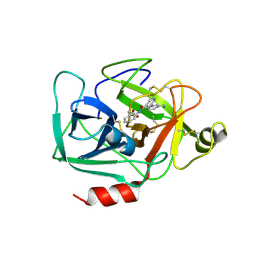 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
5B3J
 
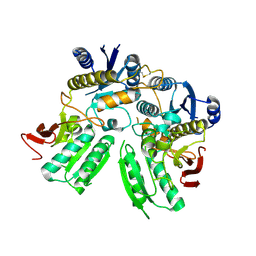 | | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil | | Descriptor: | Fab, heavy chain, light chain, ... | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil
Nature, 534, 2016
|
|
1L2K
 
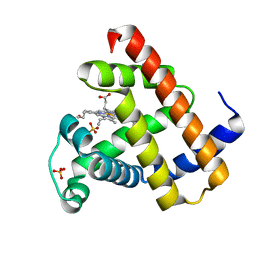 | | Neutron Structure Determination of Sperm Whale Met-Myoglobin at 1.5A Resolution. | | Descriptor: | AMMONIUM CATION WITH D, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Tanaka, I, Engler, N, Niimura, N, Parak, F.G. | | Deposit date: | 2002-02-21 | | Release date: | 2002-08-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Hydrogen and deuterium in myoglobin as seen by a neutron structure determination at 1.5 A resolution.
Biophys.Chem., 95, 2002
|
|
4LS5
 
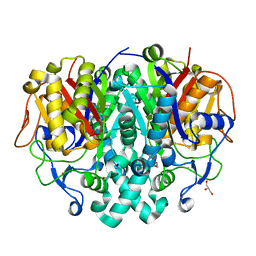 | |
6AWC
 
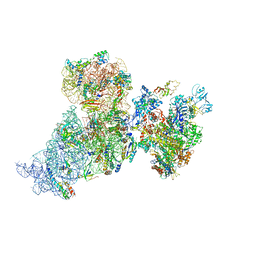 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
4MHU
 
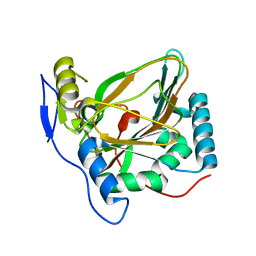 | | Crystal structure of EctD from S. alaskensis with bound Fe | | Descriptor: | Ectoine hydroxylase, FE (III) ION, N-DODECYL-N,N-DIMETHYLGLYCINATE | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4M59
 
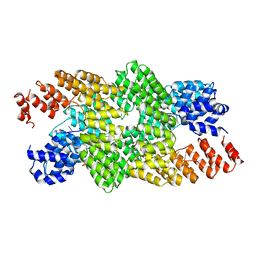 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
7OS4
 
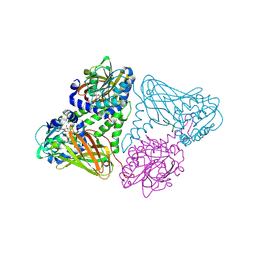 | | Crystal structure of mouse CARM1 in complex with histone H3_13-31 K18 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.1, Histone-arginine methyltransferase CARM1 | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
4LT7
 
 | | Crystal structure of the c2a domain of rabphilin-3a in complex with a calcium | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Verdaguer, N, Ferrer-Orta, C, Buxaderas, M, Corbalan-Garcia, S, Perez-Sanchez, D, Guerrero-Valero, M, Luengo, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Guillen, J. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7OKP
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4MHR
 
 | | Crystal structure of EctD from S. alaskensis in its apoform | | Descriptor: | Ectoine hydroxylase | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
