3DL4
 
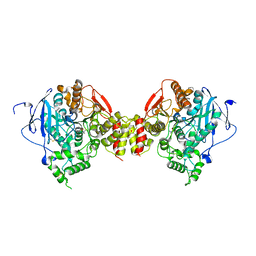 | | Non-Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | Acetylcholinesterase, HEXAETHYLENE GLYCOL | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
1CZ8
 
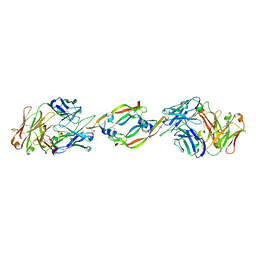 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH AN AFFINITY MATURED ANTIBODY | | Descriptor: | HEAVY CHAIN OF NEUTRALIZING ANTIBODY, LIGHT CHAIN OF NEUTRALIZING ANTIBODY, SULFATE ION, ... | | Authors: | Chen, Y, Wiesmann, C, Fuh, G, Li, B, Christinger, H.W, McKay, P, de Vos, A.M, Lowman, H.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured Fab in complex with antigen.
J.Mol.Biol., 293, 1999
|
|
3R5T
 
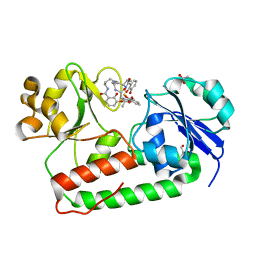 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3R5S
 
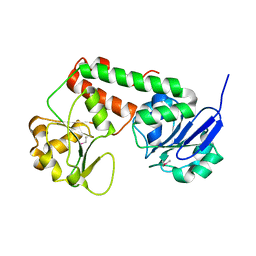 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
8K5Q
 
 | | Crystal structure of YajQ STM0435 with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), YajQ | | Authors: | Dai, Y, Zhang, M, Wang, W, Li, B. | | Deposit date: | 2023-07-23 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A c-di-GMP binding effector STM0435 modulates flagellar motility and pathogenicity in Salmonella
Virulence, 15, 2024
|
|
8K4I
 
 | |
8BV1
 
 | | Peptide inhibitor P4 in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, P4 peptide inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
8CJ3
 
 | | Urea-based foldamer inhibitor c3u_7 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_7 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ1
 
 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ2
 
 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
5H5P
 
 | |
5HKP
 
 | | Crystal structure of mouse Tankyrase/human TRF1 complex | | Descriptor: | Tankyrase-1, Telomeric repeat-binding factor 1 | | Authors: | Wang, Z, Li, B, Rao, Z, Xu, W. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a tankyrase 1-telomere repeat factor 1 complex.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
2VFX
 
 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
7M1N
 
 | |
7M1O
 
 | |
6UFR
 
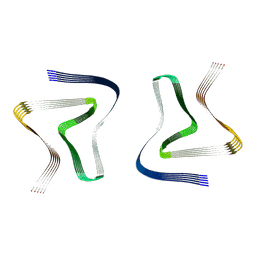 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4REY
 
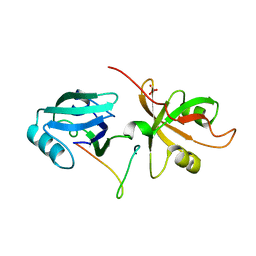 | | Crystal Structure of the GRASP65-GM130 C-terminal peptide complex | | Descriptor: | Golgi reassembly-stacking protein 1, Golgin subfamily A member 2, SULFATE ION | | Authors: | Shi, N, Hu, F, Li, B. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Interaction between the Golgi Reassembly-stacking Protein GRASP65 and the Golgi Matrix Protein GM130.
J.Biol.Chem., 290, 2015
|
|
7XXI
 
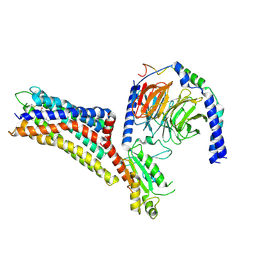 | | Cryo-EM structure of the purinergic receptor P2Y12R in complex with 2MeSADP and Gi | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
7XXH
 
 | | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(11) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
6PES
 
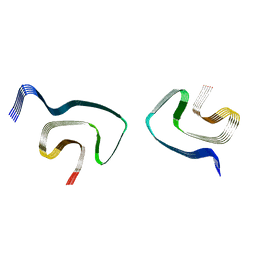 | | Cryo-EM structure of alpha-synuclein H50Q Wide Fibril | | Descriptor: | Alpha-synuclein | | Authors: | Boyer, D.R, Li, B, Sawaya, M.R, Jiang, L, Eisenberg, D.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PEO
 
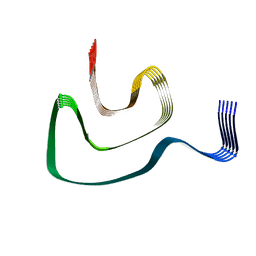 | | Cryo-EM structure of alpha-synuclein H50Q Narrow Fibril | | Descriptor: | Alpha-synuclein | | Authors: | Boyer, D.R, Li, B, Sawaya, M.R, Jiang, L, Eisenberg, D.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4BBQ
 
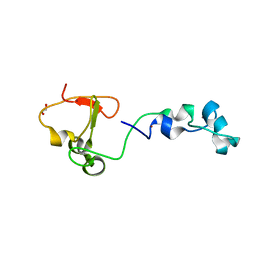 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
6IDE
 
 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
