3GD0
 
 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase) | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
4GO3
 
 | |
3U1Y
 
 | | Potent Inhibitors of LpxC for the Treatment of Gram-Negative Infections | | Descriptor: | (2R)-N-hydroxy-2-methyl-2-(methylsulfonyl)-4-{4'-[3-(morpholin-4-yl)propoxy]biphenyl-4-yl}butanamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Brown, M, Abramite, J, Liu, S. | | Deposit date: | 2011-09-30 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent inhibitors of LpxC for the treatment of Gram-negative infections.
J.Med.Chem., 55, 2012
|
|
7DTW
 
 | | Human Calcium-Sensing Receptor in the inactive close-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTU
 
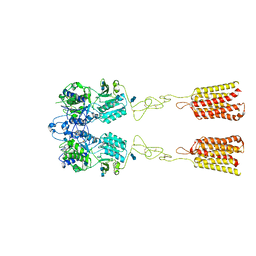 | | Human Calcium-Sensing Receptor bound with L-Trp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTT
 
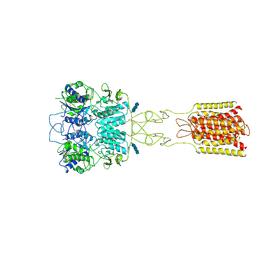 | | Human Calcium-Sensing Receptor bound with calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Sun, D.M, Liu, L, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTV
 
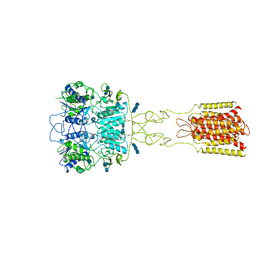 | | Human Calcium-Sensing Receptor bound with L-Trp and calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
2KWF
 
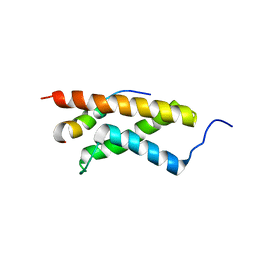 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
3UHM
 
 | |
8HPV
 
 | |
8HP9
 
 | |
8HPQ
 
 | |
2JJ3
 
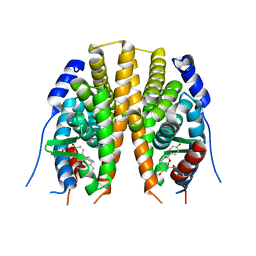 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7CFT
 
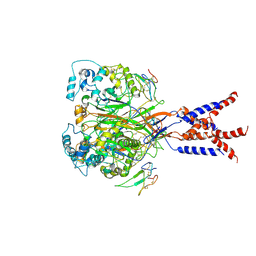 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFS
 
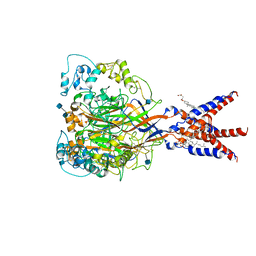 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
2PP4
 
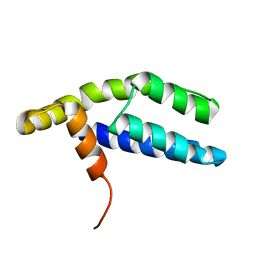 | | Solution Structure of ETO-TAFH refined in explicit solvent | | Descriptor: | Protein ETO | | Authors: | Wei, Y, Liu, S, Lausen, J, Woodrell, C, Cho, S, Biris, N, Kobayashi, N, Yokoyama, S, Werner, M.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription
Nat.Struct.Mol.Biol., 14, 2007
|
|
7Z5S
 
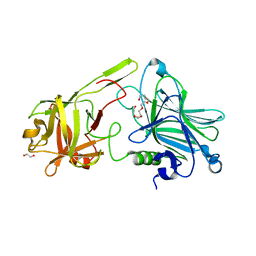 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin, HEXAETHYLENE GLYCOL, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
7Z5T
 
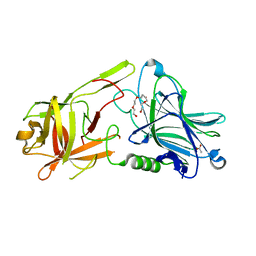 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain | | Descriptor: | ACETATE ION, Botulinum neurotoxin, PENTAETHYLENE GLYCOL | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
8AGK
 
 | | Botulinum neurotoxin subtype A6 cell binding domain in complex with GD1a ganglioside | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Newell, A.R, Mojanaga, O.O. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8ALP
 
 | | Botulinum neurotoxin A6 cell binding domain crystal form II | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-08-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8IFG
 
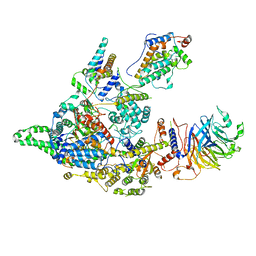 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
4OHD
 
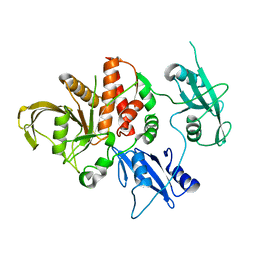 | | LEOPARD Syndrome-Associated SHP2/A461T mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
4OHL
 
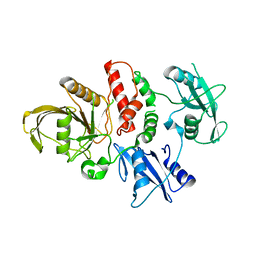 | | LEOPARD Syndrome-Associated SHP2/T468M mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
8HPU
 
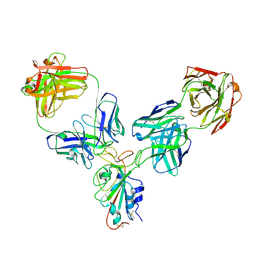 | |
8HQ7
 
 | |
