6L08
 
 | |
8I79
 
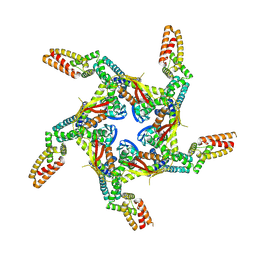 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
6PBJ
 
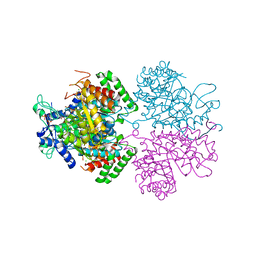 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase with Gly190Pro mutation | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Jiao, W, Fan, Y, Blackmore, N.J, Parker, E.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A single amino acid substitution uncouples catalysis and allostery in an essential biosynthetic enzyme in Mycobacterium tuberculosis .
J.Biol.Chem., 295, 2020
|
|
8JZL
 
 | |
8JZO
 
 | |
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | CryoEM structure of the NADP-dependent malic enzyme in complex with oxaloacetate
To Be Published
|
|
8K0A
 
 | |
5XBO
 
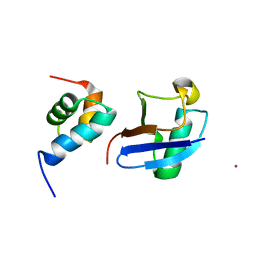 | | Lanthanoid tagging via an unnatural amino acid for protein structure characterization | | Descriptor: | Polyubiquitin-B, TERBIUM(III) ION, UV excision repair protein RAD23 homolog A | | Authors: | Jiang, W, Gu, X, Dong, X, Tang, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Lanthanoid tagging via an unnatural amino acid for protein structure characterization
J. Biomol. NMR, 67, 2017
|
|
8K03
 
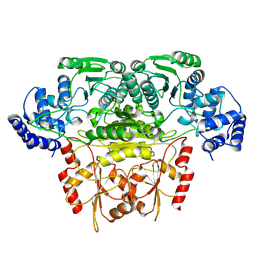 | |
3RZI
 
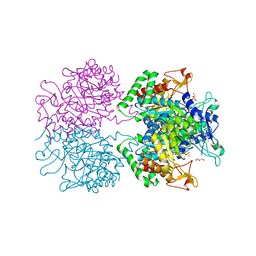 | | The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis cocrystallized and complexed with phenylalanine and tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jiao, W, Jameson, G.B, Hutton, R.D, Parker, E.J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dynamic cross-talk among remote binding sites: the molecular basis for unusual synergistic allostery.
J.Mol.Biol., 415, 2012
|
|
7YGI
 
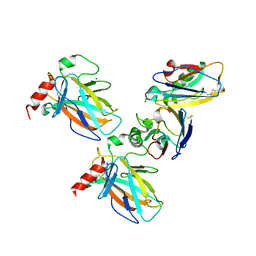 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
7XJL
 
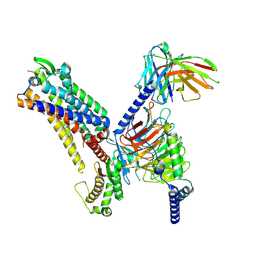 | | Cryo-EM structure of the spexin-bound GALR2-miniGq complex | | Descriptor: | Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XJK
 
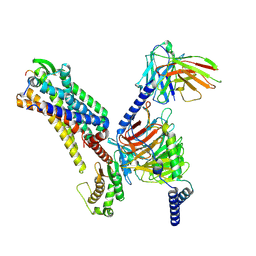 | |
7XJJ
 
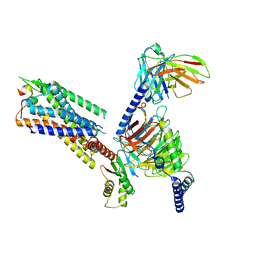 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1VKT
 
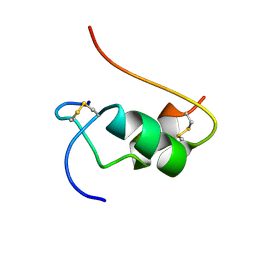 | | HUMAN INSULIN TWO DISULFIDE MODEL, NMR, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Frank, B.H, Jia, W.H, Chu, Y.C, Wang, S.H, Burke, G.T, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1996-10-14 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue.
J.Mol.Biol., 264, 1996
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1J73
 
 | | Crystal structure of an unstable insulin analog with native activity. | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Wan, Z, Zhao, M, Nakagawa, S, Jia, W, Weiss, M.A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1SJU
 
 | | MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
5CYA
 
 | | Crystal structure of Arl2 GTPase-activating protein tubulin cofactor C (TBCC) | | Descriptor: | SULFATE ION, Tubulin-specific chaperone C | | Authors: | Nithianantham, S, Le, S, Seto, E, Jia, W, Leary, J, Corbett, K.D, Moore, J.K, Al-Bassam, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alpha beta-tubulin pool for microtubule dynamics.
Elife, 4, 2015
|
|
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1SJT
 
 | | MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
3P33
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3P2X
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
