6NO8
 
 | |
6NOK
 
 | | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B1 protein loaded with myristic acid (C14:0) to 1.69 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, MYRISTIC ACID, SODIUM ION | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B1 protein loaded with myristic acid (C14:0) to 1.69 Angstrom resolution
To Be Published
|
|
6O0J
 
 | | M.tb MenD with ThDP and Inhibitor bound | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ACETATE ION, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Ho, N.A.T, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6O0N
 
 | | M.tb MenD with Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Ho, N.A.T, Bashiri, G, Bulloch, E.M, Nigon, L.V, Jirgis, E.M.N, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6OEC
 
 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | Descriptor: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
7S15
 
 | | GLP-1 receptor bound with Pfizer small molecule agonist | | Descriptor: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Liu, Y, Dias, J.M, Han, S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
4CKY
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | 2,2-dimethyl-3-dehydroquinic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
7S81
 
 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
5JHQ
 
 | |
7S6H
 
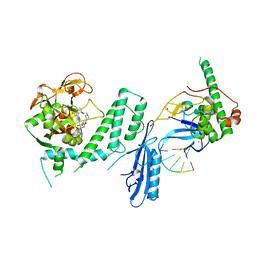 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
5JJK
 
 | | Rho transcription termination factor bound to rA7 and 6 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JES
 
 | | Human carbonic anhydrase II (V121T) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.205 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7S6M
 
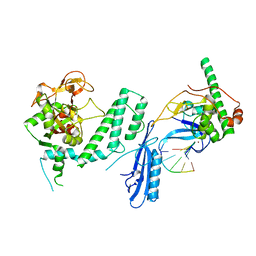 | |
7S68
 
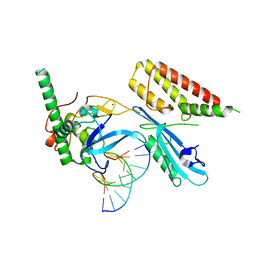 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
4CXA
 
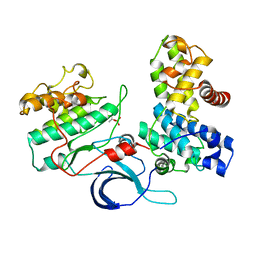 | | Crystal structure of the human CDK12-cyclin K complex bound to AMPPNP | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Nowak, R, Goubin, S, Mahajan, R.P, Kopec, J, Froese, S, Tallant, C, Carpenter, E.P, Mackenzie, A, Faust, B, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
5IUW
 
 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ and IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
7RZI
 
 | | Insulin Degrading Enzyme pC/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
 | | Insulin Degrading Enzyme O/O | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
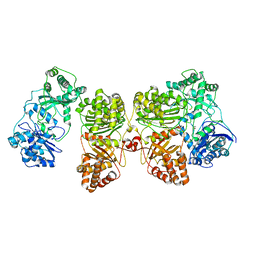 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
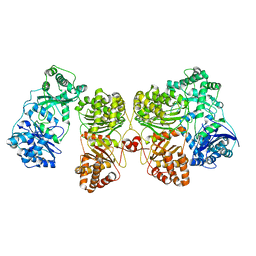 | | Insulin Degrading Enzyme O/pO | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
4CKZ
 
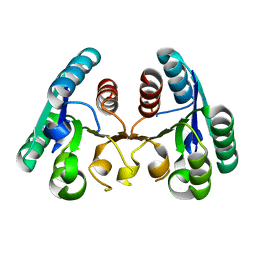 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
7S51
 
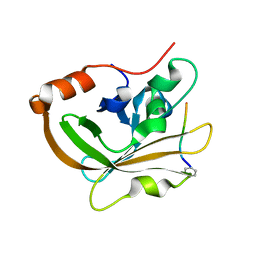 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | | Descriptor: | LEU-PRO-ALA-THR-ALA, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
