8HCN
 
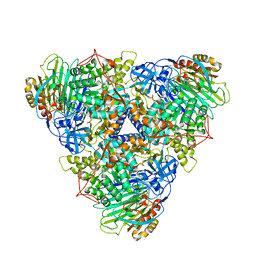 | | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | | Descriptor: | Urease accessory protein UreD, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
5EDD
 
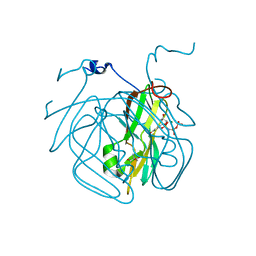 | | Crystal structure of Mycobacterium tuberculosis dUTPase R140K, H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Lopata, A, Harmat, V, Toth, J, Vertessy, G.B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
4V0B
 
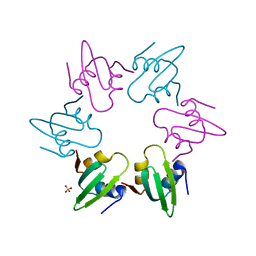 | | Escherichia coli FtsH hexameric N-domain | | Descriptor: | ATP-DEPENDENT ZINC METALLOPROTEASE FTSH, SULFATE ION | | Authors: | Serek-Heuberger, J, Martin, J, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Evolution of N-Domains in Aaa Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
4V06
 
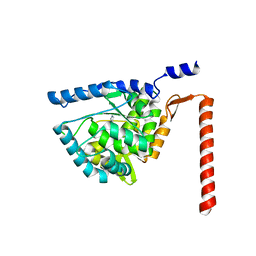 | | Crystal structure of human tryptophan hydroxylase 2 (TPH2), catalytic domain | | Descriptor: | FE (III) ION, IMIDAZOLE, TRYPTOPHAN 5-HYDROXYLASE 2 | | Authors: | Kopec, J, Oberholzer, A, Fitzpatrick, F, Newman, J, Tallant, C, Kiyani, W, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structure of Human Tryptophane Hydroxylase 2 (Tph2), Catalytic Domain
To be Published
|
|
5L2K
 
 | | Crystal structure of GEM42 TCR-CD1b-GMM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, Beta-2-microglobulin, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
4V0Q
 
 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
1I70
 
 | | CRYSTAL STRUCTURE OF RNASE SA Y86F MUTANT | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Urbanikova, L. | | Deposit date: | 2001-03-07 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine hydrogen bonds make a large contribution to protein stability.
J.Mol.Biol., 312, 2001
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5LA6
 
 | | Tubulin-pironetin complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Setter, J, Waight, A.B, Bargsten, K, Murga, J, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pironetin Binds Covalently to alpha Cys316 and Perturbs a Major Loop and Helix of alpha-Tubulin to Inhibit Microtubule Formation.
J.Mol.Biol., 428, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
1HNI
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
4USD
 
 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6L7P
 
 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
8SBB
 
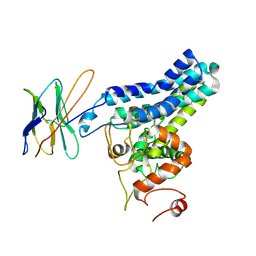 | | Cryo-EM structure of FtAlkB | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION, ... | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and mechanism of the alkane-oxidizing enzyme AlkB.
Nat Commun, 14, 2023
|
|
6QN8
 
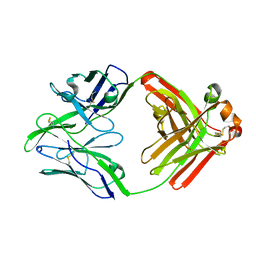 | | Structure of bovine anti-RSV Fab B13 | | Descriptor: | CHLORIDE ION, Heavy chain of bovine anti-RSV B13 Fab, Light chain of bovine anti-RSV Fab B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QSX
 
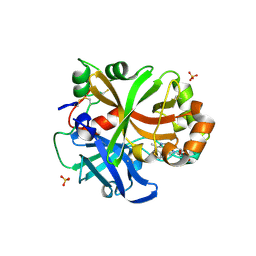 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8S99
 
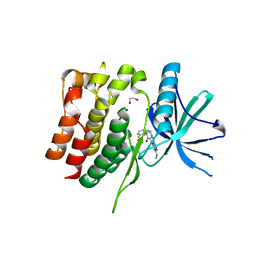 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 11 | | Descriptor: | (8S)-N-[(1R,2S)-2-fluorocyclopropyl]-5-{[(1M,2'M)-3'-fluoro-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
6DHI
 
 | |
8S9A
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with TAK-279 | | Descriptor: | (8S)-N-[(1R,2R)-2-methoxycyclobutyl]-7-(methylamino)-5-{[(1P,2'P)-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S98
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 8 | | Descriptor: | (8S)-N-cyclopropyl-5-[(2-methoxypyridin-3-yl)amino]-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S9B
 
 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
