7MHX
 
 | | KcsA E71V closed gate with Ba2+ | | Descriptor: | BARIUM ION, DIACYL GLYCEROL, Fab heavy chain, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7MUB
 
 | | KcsA Open gate E71V mutant in Potassium | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7MJT
 
 | | KcsA open gate E71V mutant with Barium | | Descriptor: | BARIUM ION, Fab heavy chain, Fab light chain, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7MK6
 
 | | KcsA open gate E71V mutant with sodium | | Descriptor: | Fab heavy chain, Fab light chain, pH-gated potassium channel KcsA | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
4ZUV
 
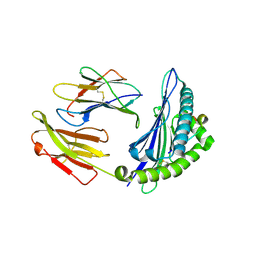 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Env-RW12 | | Descriptor: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUU
 
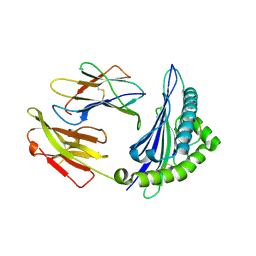 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-CF9 | | Descriptor: | Beta-2-microglobulin, CYS-THR-SER-GLU-GLU-MET-ASN-ALA-PHE, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
6I3L
 
 | | Bilirubin oxidase from Myrothecium verrucaria, mutant W396F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Koval, T, Svecova, L, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
7YZ4
 
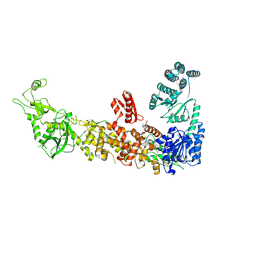 | | Mouse endoribonuclease Dicer (composite structure) | | Descriptor: | Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7YYM
 
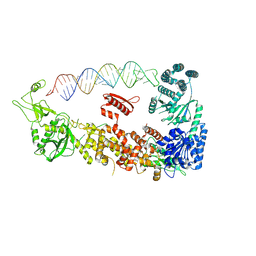 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7YYN
 
 | | Mammalian Dicer in the dicing state with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Isoform 2 of Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
7MMD
 
 | |
1S1T
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
3I3T
 
 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3IF7
 
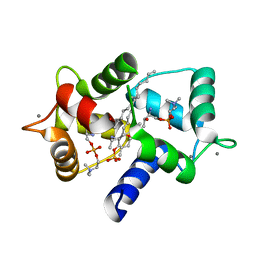 | | Structure of Calmodulin complexed with its first endogenous inhibitor, sphingosylphosphorylcholine | | Descriptor: | 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, CALCIUM ION, Calmodulin | | Authors: | Kovacs, E, Harmat, V, Toth, J, Vertessy, B.G, Modos, K, Kardos, J, Liliom, K. | | Deposit date: | 2009-07-24 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of calmodulin binding to a signaling sphingolipid reveal new aspects of lipid-protein interactions
Faseb J., 24, 2010
|
|
7MX9
 
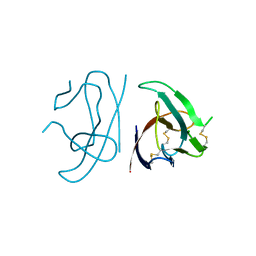 | |
5LNL
 
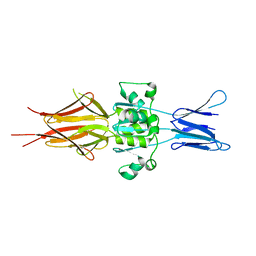 | | Crystal structure of Hsf 1608-1749 putative domain 1 | | Descriptor: | Hsf | | Authors: | Thomsen, M, Wright, J, Ridley, J, Goldman, A. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of PD1, a Haemophilus surface fibril domain.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2C47
 
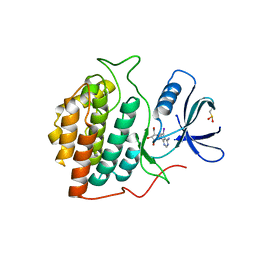 | | Structure of casein kinase 1 gamma 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, CASEIN KINASE 1 GAMMA 2 ISOFORM, MAGNESIUM ION | | Authors: | Bunkoczi, G, Rellos, P, Das, S, Ugochukwu, E, Fedorov, O, Sobott, F, Eswaran, J, Amos, A, Ball, L, von Delft, F, Bullock, A, Debreczeni, J, Turnbull, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-16 | | Release date: | 2005-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Casein Kinase 1 Gamma 2
To be Published
|
|
7Z7W
 
 | | REP-related Chom18 variant with double GC base pairing | | Descriptor: | Chom18-GC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z82
 
 | | REP-related Chom18 variant with double AG mismatch | | Descriptor: | Chom18-AG DNA, STRONTIUM ION | | Authors: | Svoboda, J, Kolenko, P, Berdar, D, Schneider, B. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
8OVX
 
 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
7Z7Z
 
 | | REP-related Chom18 variant with double TA base pair | | Descriptor: | Chom18-TA DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
