1UAT
 
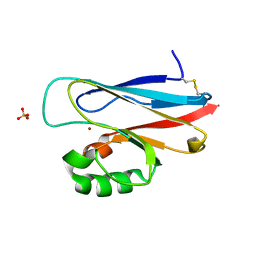 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | Descriptor: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | Authors: | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | Deposit date: | 2003-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
8GB0
 
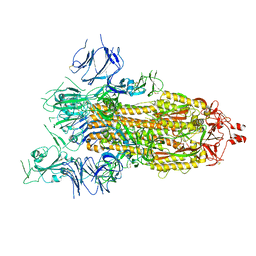 | |
8G57
 
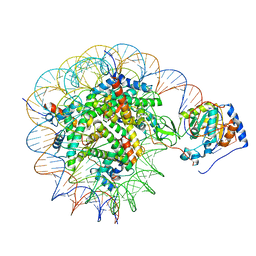 | | Structure of nucleosome-bound Sirtuin 6 deacetylase | | Descriptor: | DNA strand 1, DNA strand 2, Histone H2A type 1-B/E, ... | | Authors: | Chio, U.S, Rechiche, O, Bryll, A.R, Zhu, J, Feldman, J.L, Peterson, C.L, Tan, S, Armache, J.-P. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of the human Sirtuin 6-nucleosome complex.
Sci Adv, 9, 2023
|
|
4IDI
 
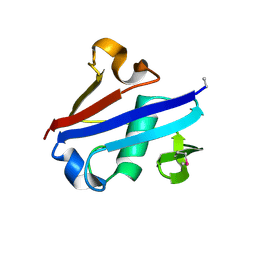 | | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420 | | Descriptor: | GLYCEROL, Oryza sativa Rurm1-related | | Authors: | Wernimont, A.K, Tempel, W, Lew, J, Walker, J, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Bountra, C, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-12 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420
To be Published
|
|
4IEL
 
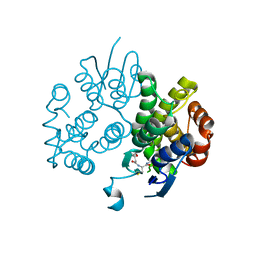 | | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione
To be Published
|
|
7TA3
 
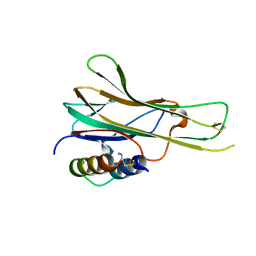 | |
4D4B
 
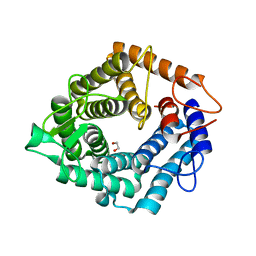 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4L
 
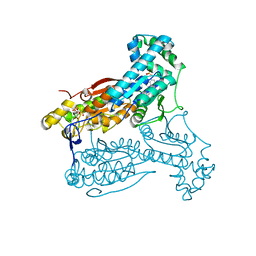 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4CRT
 
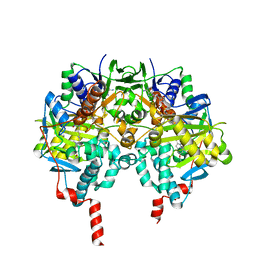 | | Crystal structure of human monoamine oxidase B in complex with the multi-target inhibitor ASS234 | | Descriptor: | (E)-N-methyl-N-[[1-methyl-5-[3-[1-(phenylmethyl)piperidin-4-yl]propoxy]indol-2-yl]methyl]prop-1-en-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Esteban, G, Allan, J, Samadi, A, Mattevi, A, Unzeta, M, Marco-Contelles, J, Binda, C, Ramsay, R.R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-04-02 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Analysis of the Irreversible Inhibition of Human Monoamine Oxidases by Ass234, a Multi-Target Compound Designed for Use in Alzheimer'S Disease.
Biochim.Biophys.Acta, 1844, 2014
|
|
7TCI
 
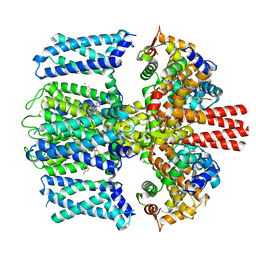 | | Structure of Xenopus KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, CALCIUM ION, Calmodulin-1, ... | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
6SDX
 
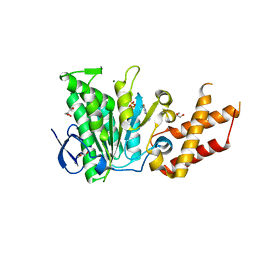 | | Salmonella ATPase InvC with ATP gamma S | | Descriptor: | ATP synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bernal, I, Roemermann, J, Flacht, L, Lunelli, M, Uetrecht, C, Kolbe, M. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural analysis of ligand-bound states of the Salmonella type III secretion system ATPase InvC.
Protein Sci., 28, 2019
|
|
6SY1
 
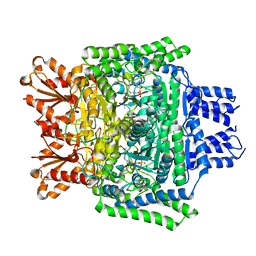 | | Crystal structure of the human 2-oxoadipate dehydrogenase DHTKD1 (E1) | | Descriptor: | MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Shrestha, L, Pena, I.A, Coker, J, Kolker, S, Nicola, B.B, von Delft, F, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism.
Iucrj, 7, 2020
|
|
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
3R2B
 
 | | MK2 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, van Zeeland, M, Versteegh, J. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4IFP
 
 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
8G4G
 
 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
4IHC
 
 | | Crystal structure of probable mannonate dehydratase Dd703_0947 (target EFI-502222) from Dickeya dadantii Ech703 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mannonate dehydratase Dd703_0947 from Dickeya dadantii Ech703
To be Published
|
|
7TCP
 
 | | Structure of Xenopus KCNQ1-CaM | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
4IKB
 
 | | Crystal structure of SNX11 PX domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Xu, J, Xu, T, Liu, J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Sorting Nexin 11 (SNX11) Reveals a Novel Extended PX Domain (PXe Domain) Critical for the Inhibition of Sorting Nexin 10 (SNX10) Induced Vacuolation
to be published
|
|
3EQR
 
 | | Crystal Structure of Ack1 with compound T74 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N~3~-(2,6-dimethylphenyl)-1-(3-methoxy-3-methylbutyl)-N~6~-(4-piperazin-1-ylphenyl)-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8JJE
 
 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
6SQK
 
 | |
4BZ1
 
 | | Structure of dengue virus EDIII in complex with Fab 3e31 | | Descriptor: | CHLORIDE ION, ENVELOPE PROTEIN, FAB 3E31 HEAVY CHAIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 3E31
To be Published
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
8G8W
 
 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
