3BLA
 
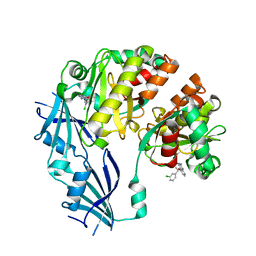 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
4QKJ
 
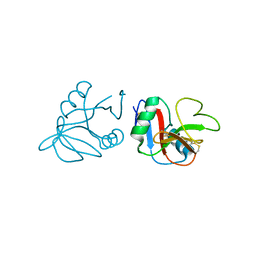 | | Glycosylated form of human LLT1, a ligand for NKR-P1, in this structure forming hexamers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D | | Authors: | Skalova, T, Blaha, J, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5D7D
 
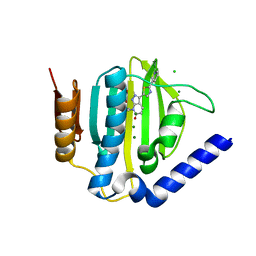 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
3BL9
 
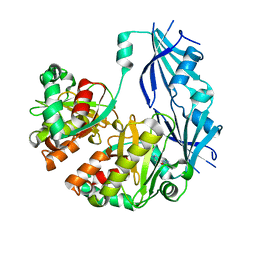 | | Synthetic Gene Encoded DcpS bound to inhibitor DG157493 | | Descriptor: | 5-{[1-(2,3-dichlorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
2GHI
 
 | | Crystal Structure of Plasmodium yoelii Multidrug Resistance Protein 2 | | Descriptor: | SULFATE ION, transport protein | | Authors: | Dong, A, Gao, M, Choe, J, Zhao, Y, Lew, J, Wasney, G, Alam, Z, Melone, M, Kozieradzki, I, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3BL7
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG156844 | | Descriptor: | 5-{[1-(2-fluorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3CCK
 
 | | Human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69 | | Authors: | Brynda, J, Vanek, O, Rezacova, P. | | Deposit date: | 2008-02-26 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Soluble recombinant CD69 receptors optimized to have an exceptional physical and chemical stability display prolonged circulation and remain intact in the blood of mice
Febs J., 275, 2008
|
|
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
1SL8
 
 | | Calcium-loaded apo-aequorin from Aequorea victoria | | Descriptor: | Aequorin 1, CALCIUM ION | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
6T7U
 
 | | Carborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, Carborane inhibitor, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
8SY5
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY7
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
7P7A
 
 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P77
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P7B
 
 | | SARS-CoV-2 spike protein in complex with sybody no68 in a 1up/2down conformation | | Descriptor: | Spike glycoprotein | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
6T9Z
 
 | | Nidocarborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, Nidocarborane, ... | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
7OZV
 
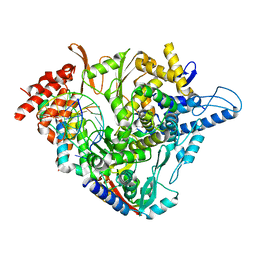 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZU
 
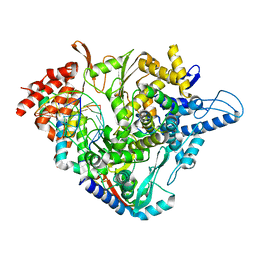 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1CCV
 
 | |
1SL7
 
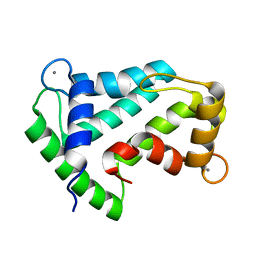 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
8HQG
 
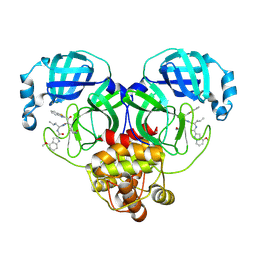 | |
