3U2Z
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
7TL7
 
 | | 1.90A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Sa-D2) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, IMIDAZOLE, SODIUM ION, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
7TL8
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from S. aureus in complex with a macrocyclic peptide inhibitor (Sa-D3) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION, Peptide Sa-D3 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
4E5D
 
 | | 2.2A resolution structure of a firefly luciferase-benzothiazole inhibitor complex | | Descriptor: | 2-(2-fluorophenyl)-6-methoxy-1,3-benzothiazole, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Throne, N, Shen, M, Auld, D.S, Inglese, J. | | Deposit date: | 2012-03-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.
Chem.Biol., 19, 2012
|
|
3ME3
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNF
 
 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
5KGN
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
3RIX
 
 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
3GR4
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-[(2,6-difluorophenyl)sulfonyl]-4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)piperazine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3GQY
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-4-[(4-methoxyphenyl)sulfonyl]piperazine, L(+)-TARTARIC ACID, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3H6O
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(2-fluorobenzyl)-2,4-dimethyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3IES
 
 | | Firefly luciferase inhibitor complex | | Descriptor: | 5'-O-[(R)-[({3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]phenyl}carbonyl)oxy](hydroxy)phosphoryl]adenosine, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IER
 
 | | Firefly luciferase apo structure (P41 form) with PEG 400 bound | | Descriptor: | Luciferin 4-monooxygenase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IEP
 
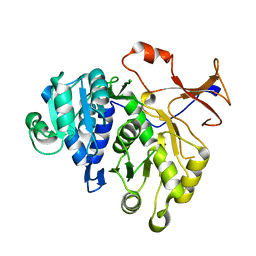 | | Firefly luciferase apo structure (P41 form) | | Descriptor: | Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
