3TVJ
 
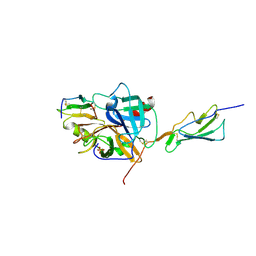 | | Catalytic fragment of MASP-2 in complex with its specific inhibitor developed by directed evolution on SGCI scaffold | | Descriptor: | Mannan-binding lectin serine protease 2 A chain, Mannan-binding lectin serine protease 2 B chain, Protease inhibitor SGPI-2, ... | | Authors: | Heja, D, Harmat, V, Dobo, J, Szasz, R, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2011-09-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
3GOV
 
 | | Crystal structure of the catalytic region of human MASP-1 | | Descriptor: | GLYCEROL, MASP-1 | | Authors: | Harmat, V, Dobo, J, Beinrohr, L, Sebestyen, E, Zavodszky, P, Gal, P. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MASP-1, a promiscuous complement protease: structure of its catalytic region reveals the basis of its broad specificity.
J.Immunol., 183, 2009
|
|
2QY0
 
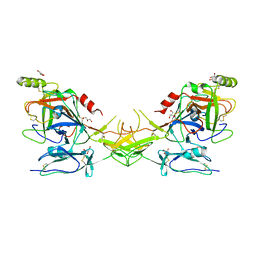 | | Active dimeric structure of the catalytic domain of C1r reveals enzyme-product like contacts | | Descriptor: | Complement C1r subcomponent, GLYCEROL | | Authors: | Kardos, J, Harmat, V, Pallo, A, Barabas, O, Szilagyi, K, Graf, L, Naray-Szabo, G, Goto, Y, Zavodszky, P, Gal, P. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Mol.Immunol., 45, 2008
|
|
2OAY
 
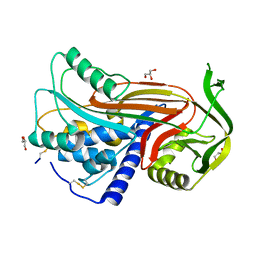 | | Crystal structure of latent human C1-inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Plasma protease C1 inhibitor | | Authors: | Harmat, V, Beinrohr, L, Gal, P, Dobo, J. | | Deposit date: | 2006-12-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease
J.Biol.Chem., 282, 2007
|
|
1Q3X
 
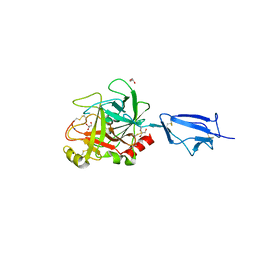 | | Crystal structure of the catalytic region of human MASP-2 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 2, SODIUM ION | | Authors: | Harmat, V, Gal, P, Kardos, J, Szilagyi, K, Ambrus, G, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions
J.Mol.Biol., 342, 2004
|
|
4DJZ
 
 | | Catalytic fragment of masp-1 in complex with its specific inhibitor developed by directed evolution on sgci scaffold | | Descriptor: | Mannan-binding lectin serine protease 1 heavy chain, Mannan-binding lectin serine protease 1 light chain, Protease inhibitor SGPI-2 | | Authors: | Heja, D, Harmat, V, Fodor, K, Wilmanns, M, Dobo, J, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
8JIC
 
 | |
8JIH
 
 | |
4WD4
 
 | | Crystal structure of human HO1 H25R | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
7ZAO
 
 | | Structure of BPP43_05035 of Brachyspira pilosicoli | | Descriptor: | MAGNESIUM ION, Sialidase (Neuraminidase) family protein-like protein | | Authors: | Rajan, A, Gallego, P, Pelaseyed, T. | | Deposit date: | 2022-03-22 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | BPP43_05035 is a Brachyspira pilosicoli cell surface adhesin that weakens the integrity of the epithelial barrier during infection
Biorxiv, 2024
|
|
2VQW
 
 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
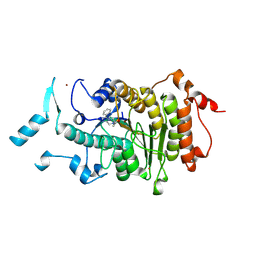 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQQ
 
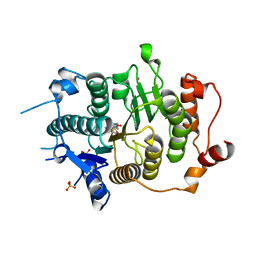 | | Structure of HDAC4 catalytic domain (a double cysteine-to-alanine mutant) bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQO
 
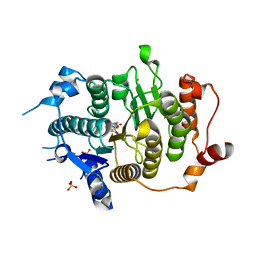 | | Structure of HDAC4 catalytic domain with a gain-of-function muation bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQJ
 
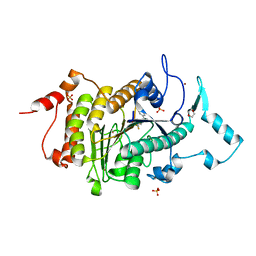 | | Structure of HDAC4 catalytic domain bound to a trifluoromethylketone inhbitor | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQV
 
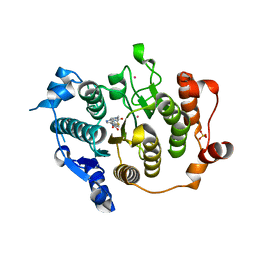 | | Structure of HDAC4 catalytic domain with a gain-of-function mutation bound to a hydroxamic acid inhibitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
8ACS
 
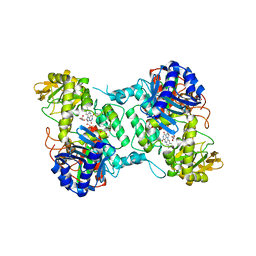 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
5FO2
 
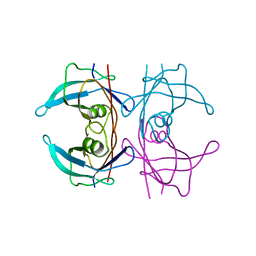 | | Structure of human transthyretin mutant A108I | | Descriptor: | TRANSTHYRETIN | | Authors: | Varejao, N, Santanna, R, Saraiva, M.J, Gallego, P, Ventura, S, Reverter, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation.
Sci Rep, 7, 2017
|
|
2V5X
 
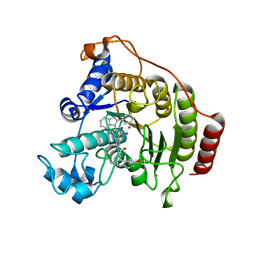 | | Crystal structure of HDAC8-inhibitor complex | | Descriptor: | (2R)-N~8~-HYDROXY-2-{[(5-METHOXY-2-METHYL-1H-INDOL-3-YL)ACETYL]AMINO}-N~1~-[2-(2-PHENYL-1H-INDOL-3-YL)ETHYL]OCTANEDIAMIDE, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C, Gallinari, P, Jones, P, Mattu, M, Carfi, A, Defrancesco, R, Steinkuhler, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
4J2T
 
 | | Inhibitor-bound Ca2+ ATPase | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9bS)-6-(acetyloxy)-3a,4-bis(butanoyloxy)-3-hydroxy-3,6,9-trimethyl-8-{[(2E)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-7-yl octanoate, PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, ... | | Authors: | Paulsen, E.S, Villadsen, J, Tenori, E, Liu, H, Lie, M.A, Bonde, D.F, Bublitz, M, Olesen, C, Autzen, H.E, Dach, I, Sehgal, P, Moller, J.V, Schiott, B, Nissen, P, Christensen, S.B. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase.
J.Med.Chem., 56, 2013
|
|
5A6I
 
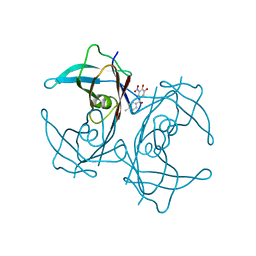 | | V122I Transthyretin structure in complex with Tolcalpone | | Descriptor: | TRANSTHYRETIN, Tolcapone | | Authors: | Reverter, D, Gallego, P, Santana, R, Ventura, S. | | Deposit date: | 2015-06-26 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Repositioning Tolcapone as a Potent Inhibitor of Transthyretin Amyloidogenesis and its Associated Cellular Toxicity
Nat.Commun., 7, 2016
|
|
3FGH
 
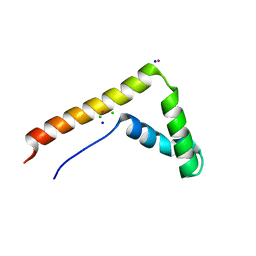 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
1MZI
 
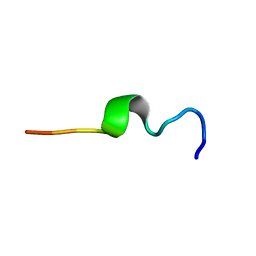 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
2BEQ
 
 | | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein | | Descriptor: | Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-11-29 | | Release date: | 2004-12-22 | | Last modified: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2BEZ
 
 | | Structure of a proteolitically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein | | Descriptor: | GLYCEROL, Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-12-02 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
