4CI2
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CI1
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | Descriptor: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (7.4 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
7B1H
 
 | |
7B1F
 
 | |
7B1J
 
 | |
6XMX
 
 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
8G46
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
8UWB
 
 | | Crystal structure of PP2A PPP2R1A-PPP2CA-PPP2R5E phosphatase. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wachter, F, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of methylation-independent PP2A assembly guides alphafold2Multimer prediction of family-wide PP2A complexes.
J.Biol.Chem., 300, 2024
|
|
8TNR
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNP
 
 | | Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40 | | Descriptor: | DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, Protein cereblon, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNQ
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TL6
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF5 | | Descriptor: | DDB1- and CUL4-associated factor 5, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1 | | Authors: | Yue, H, Hunkeler, M, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF.
Nature, 628, 2024
|
|
3GJF
 
 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Antibody heavy chain, Antibody light chain, Beta-2-microglobulin, ... | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GJE
 
 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Fab Heavy Chain, Fab Light Chain | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HAE
 
 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Antibody heavy chain, Antibody light chain, Beta-2-microglobulin, ... | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E. | | Deposit date: | 2009-05-01 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7U20
 
 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8E2D
 
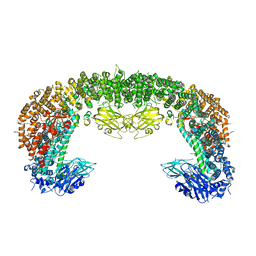 | | Cryo-EM structure of BIRC6 (consensus) | | Descriptor: | Baculoviral IAP repeat-containing protein 6 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2G
 
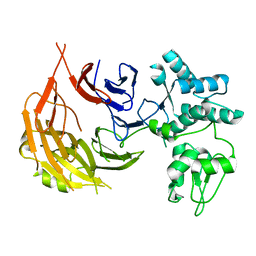 | |
8E2J
 
 | | Cryo-EM structure of BIRC6/Smac (from local refinement 1) | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Diablo IAP-binding mitochondrial protein | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2E
 
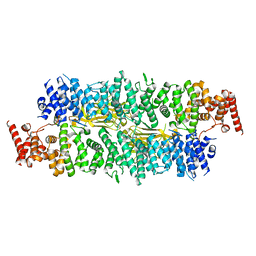 | |
8E2I
 
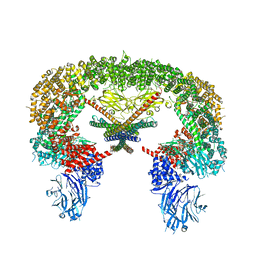 | | Cryo-EM structure of BIRC6/Smac | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Diablo IAP-binding mitochondrial protein | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2F
 
 | |
