8CDK
 
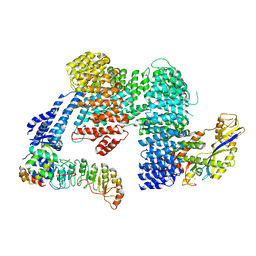 | |
8CDJ
 
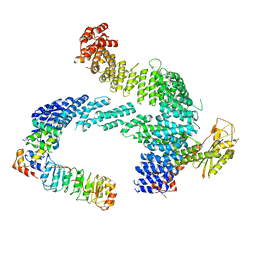 | |
7ADO
 
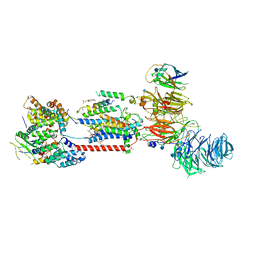 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | 著者 | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-01-20 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7ADP
 
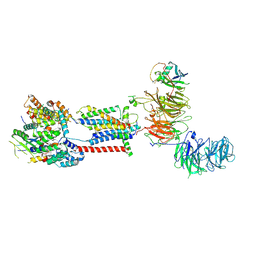 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | 著者 | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
6VJ7
 
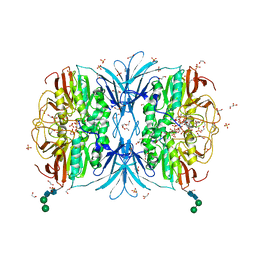 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | 登録日 | 2020-01-15 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
4RG7
 
 | |
8EBM
 
 | |
8EBL
 
 | |
8C07
 
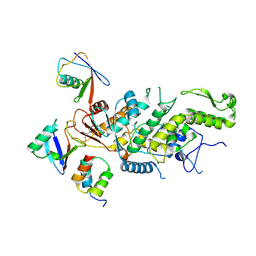 | |
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7R
 
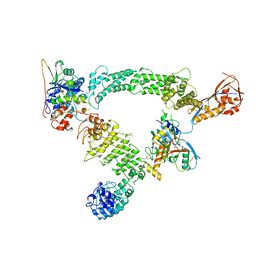 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | 分子名称: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8R5H
 
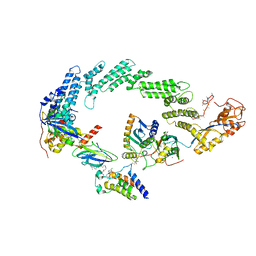 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | 分子名称: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-11-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
7ONI
 
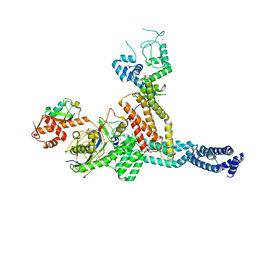 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | 分子名称: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | 著者 | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | 登録日 | 2021-05-25 | | 公開日 | 2021-09-15 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
8OIF
 
 | | Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6 | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | 著者 | Wallace, I, Kheewoong, B, Prabu, J.R, Vollrath, R, von Gronau, S, Schulman, B.A, Swatek, K.N. | | 登録日 | 2023-03-22 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Insights into the ISG15 transfer cascade by the UBE1L activating enzyme.
Nat Commun, 14, 2023
|
|
7Q51
 
 | | yeast Gid10 bound to a Phe/N-peptide | | 分子名称: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
 | | human Gid4 bound to a Phe/N-peptide | | 分子名称: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | 著者 | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
8PD0
 
 | | cryo-EM structure of Doa10 in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-11 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PDA
 
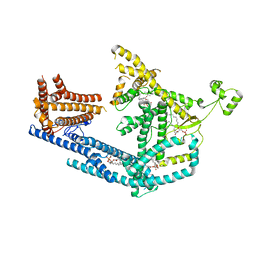 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-12 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
7WUG
 
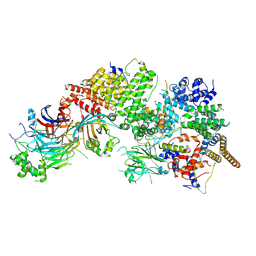 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | 分子名称: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Cheng, J.D, Schulman, B.A. | | 登録日 | 2022-02-08 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
7QQY
 
 | |
6E5A
 
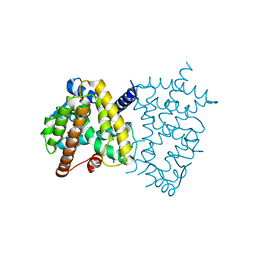 | | PPARg in complex with compound 4b | | 分子名称: | (5Z)-5-({4-[(prop-2-yn-1-yl)oxy]phenyl}methylidene)-2-sulfanylidene-1,3-thiazolidin-4-one, Peroxisome proliferator-activated receptor gamma | | 著者 | Bruning, J.B, Chua, B.S.K. | | 登録日 | 2018-07-19 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Shooting three inflammatory targets with a single bullet: Novel multi-targeting anti-inflammatory glitazones.
Eur J Med Chem, 167, 2019
|
|
6S9O
 
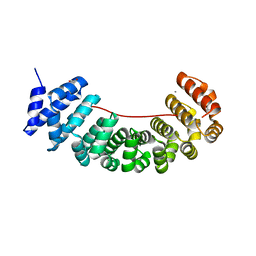 | | Designed Armadillo Repeat protein internal Lock1 fused to target peptide KRKRKLKFKR | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, designed Armadillo repeat protein with internal Lock1 fused to target peptide KRKRKLKFKR | | 著者 | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6S9M
 
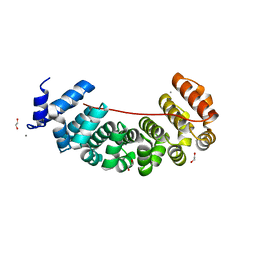 | | Designed Armadillo Repeat protein Lock2 fused to target peptide KRKRKAKITW | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Lock2_KRKRKAKITW, ... | | 著者 | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
5V83
 
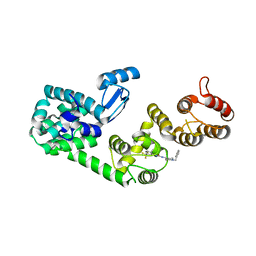 | | Structure of DCN1 bound to NAcM-HIT | | 分子名称: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | 分子名称: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
