3J9L
 
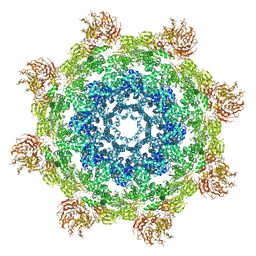 | | Structure of Dark apoptosome from Drosophila melanogaster | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HM4
 
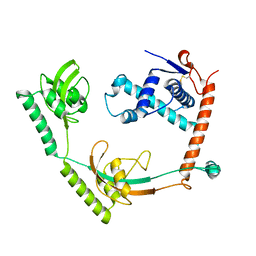 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
1IEH
 
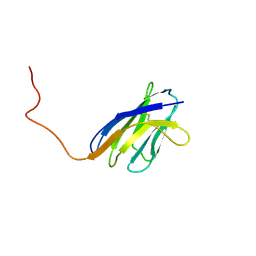 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
7BWT
 
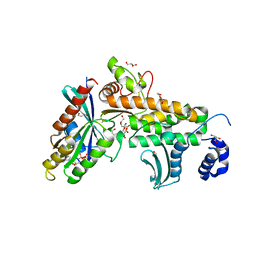 | | SopD-Rab8 complex structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jiang, K, Tong, M, Chen, Z, Gao, X. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Salmonella effector protein SopD targets Rab8 to positively and negatively modulate the inflammatory response.
Nat Microbiol, 6, 2021
|
|
6JFW
 
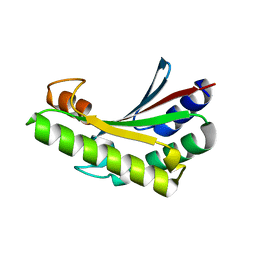 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8HM2
 
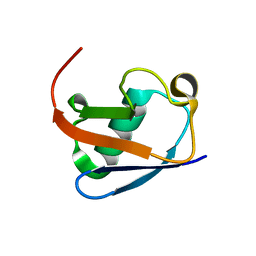 | |
8HM1
 
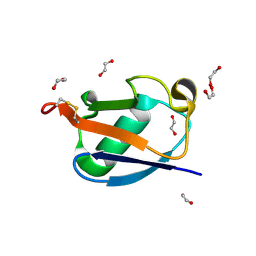 | |
6K1P
 
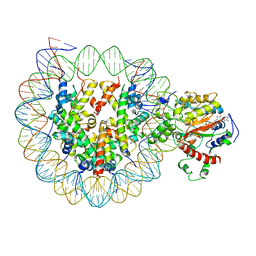 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
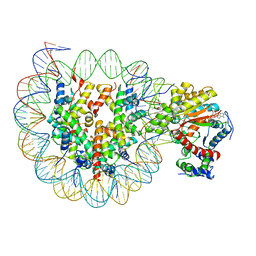 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7QZQ
 
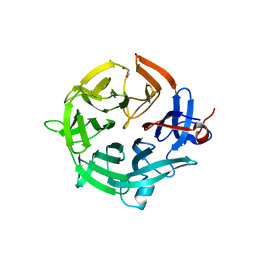 | | Crystal structure of the kelch domain of human KBTBD12 | | Descriptor: | 1,2-ETHANEDIOL, Kelch repeat and BTB domain-containing protein 12, SODIUM ION | | Authors: | Manning, C.E, Chen, Z, Chen, X, Bradshaw, W.J, Bakshi, S, Mckinley, G, Chalk, R, Burgess-Brown, N, von Delft, F, Bullock, A.N. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the kelch domain of human KBTBD12
To Be Published
|
|
4XOI
 
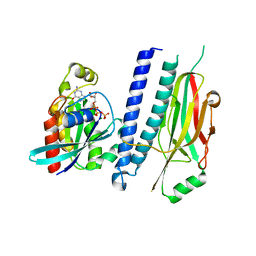 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
6EGC
 
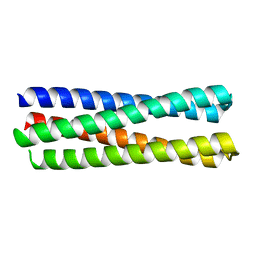 | |
8DT3
 
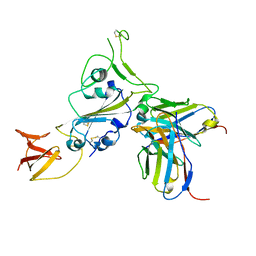 | | Cryo-EM structure of spike binding to Fab of neutralizing antibody (locally refined) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fab of SW186, Light chain Fab of SW186, ... | | Authors: | Sun, P.C, Fang, Y, Bai, X.C, Chen, Z.J. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An antibody that neutralizes SARS-CoV-1 and SARS-CoV-2 by binding to a conserved spike epitope outside the receptor binding motif.
Sci Immunol, 7, 2022
|
|
8HNV
 
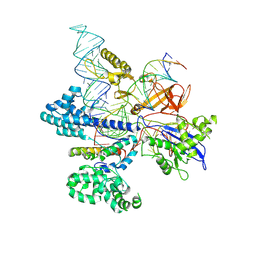 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | Descriptor: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1TQF
 
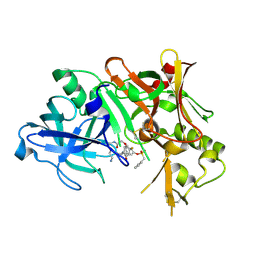 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
4XH3
 
 | |
4LZX
 
 | | Complex of IQCG and Ca2+-free CaM | | Descriptor: | Calmodulin, IQ domain-containing protein G, SULFATE ION | | Authors: | Liang, W.X, Chen, L.T, Chen, Z, Chen, S.J, Chen, S. | | Deposit date: | 2013-08-01 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and molecular features of the calmodulin-interacting protein IQCG required for haematopoiesis in zebrafish
Nat Commun, 5, 2014
|
|
7RNN
 
 | | Human ASIC1a-Nb.C1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Nanobodies Nb.C1 | | Authors: | Wu, Y, Chen, Z, Sigworth, F.J, Canessa, C.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure and analysis of nanobody binding to the human ASIC1a ion channel.
Elife, 10, 2021
|
|
2X87
 
 | | Crystal Structure of the reconstituted CotA | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Bento, I, Silva, C.S, Chen, Z, Martins, L.O, Lindley, P.F, Soares, C.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanisms Underlying Dioxygen Reduction in Laccases. Structural and Modelling Studies Focusing on Proton Transfer.
Bmc Struct.Biol., 10, 2010
|
|
2WSD
 
 | | Proximal mutations at the type 1 Cu site of CotA-laccase: I494A mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Durao, P, Chen, Z, Soares, C.M, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal Mutations at the Type 1 Copper Site of Cota Laccase: Spectroscopic, Redox, Kinetic and Structural Characterization of I494A and L386A Mutants.
Biochem.J., 412, 2008
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
4M1L
 
 | | Complex of IQCG and Ca2+-bound CaM | | Descriptor: | CALCIUM ION, Calmodulin, IQ domain-containing protein G, ... | | Authors: | Liang, W.X, Chen, L.T, Chen, Z, Chen, S.J, Chen, S. | | Deposit date: | 2013-08-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and molecular features of the calmodulin-interacting protein IQCG required for haematopoiesis in zebrafish
Nat Commun, 5, 2014
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
