5FTP
 
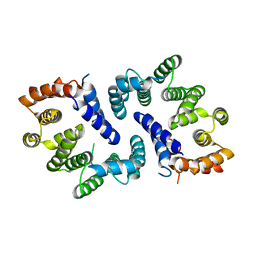 | | sulfur SAD phasing of Cdc23Nterm: data collection with a tailored X- ray beam size at 2.69 A wavelength (4.6 keV) | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8 | | Authors: | Cianci, M, Groves, M.R, Barford, D, Schneider, T.R. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Data Collection with a Tailored X-Ray Beam Size at 2.69 A Wavelength (4.6 Kev): Sulfur Sad Phasing of Cdc23Nterm
Acta Crystallogr.,Sect.D, 72, 2016
|
|
8Q84
 
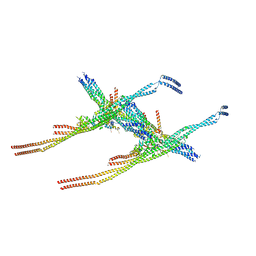 | |
2Y4I
 
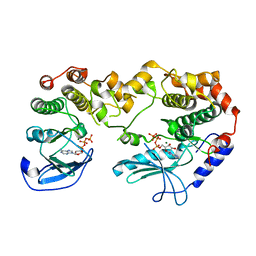 | | KSR2-MEK1 heterodimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, ... | | Authors: | Brennan, D.F, Barford, D. | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Raf-Induced Allosteric Transition of Ksr Stimulates Ksr and Raf Phosphorylation of Mek
Nature, 472, 2011
|
|
8PPR
 
 | |
6GYU
 
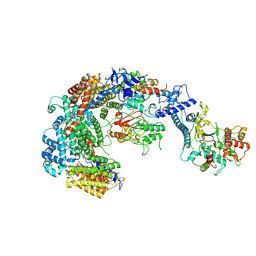 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7AAP
 
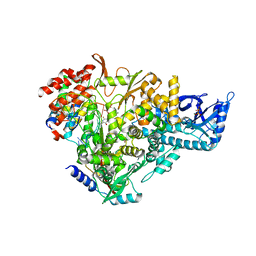 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6GYP
 
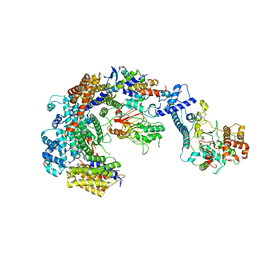 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4A2N
 
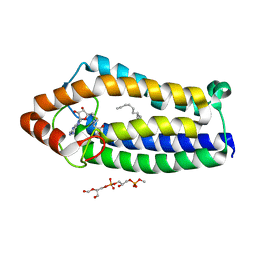 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
6TLJ
 
 | |
8QRY
 
 | |
2YIN
 
 | | STRUCTURE OF THE COMPLEX BETWEEN Dock2 AND Rac1. | | Descriptor: | DEDICATOR OF CYTOKINESIS PROTEIN 2, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 | | Authors: | Kulkarni, K.A, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple Factors Confer Specific Cdc42 and Rac Protein Activation by Dedicator of Cytokinesis (Dock) Nucleotide Exchange Factors.
J.Biol.Chem., 286, 2011
|
|
6QLD
 
 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6GYS
 
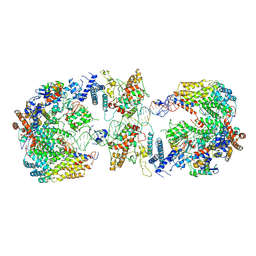 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2XPI
 
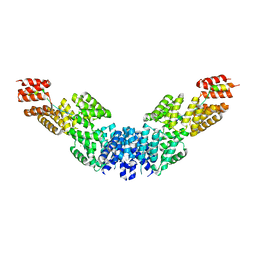 | | Crystal structure of APC/C hetero-tetramer Cut9-Hcn1 | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT CUT9, ANAPHASE-PROMOTING COMPLEX SUBUNIT HCN1 HCN1/CDC26,20S CYCLOSOME/APC COMPLEX PROTEIN HCN1, CHAPERONE-LIKE PROTEIN HCN1, ... | | Authors: | Zhang, Z, Kulkarni, K.A, Barford, D. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-30 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Apc/C Subunit Cdc16/Cut9 is a Contiguous Tetratricopeptide Superhelix with a Homo-Dimer Interface Similar to Cdc27
Embo J., 29, 2010
|
|
1RPM
 
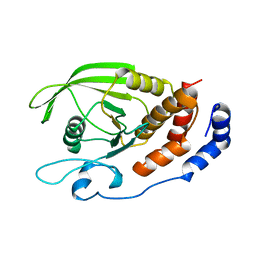 | |
4ADY
 
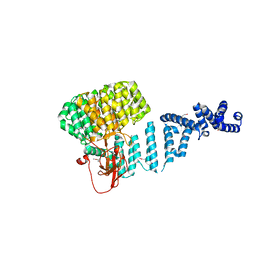 | | Crystal structure of 26S proteasome subunit Rpn2 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN2 | | Authors: | Kulkarni, K, He, J, Da Fonseca, P.C.A, Krutauz, D, Glickman, M.H, Barford, D, Morris, E.P. | | Deposit date: | 2012-01-04 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
Structure, 20, 2012
|
|
4BH6
 
 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
4CAD
 
 | | Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kulkarni, K, Manolaridis, I, Dodd, R.B, Cronin, N, Ogasawara, S, Iwata, S, Barford, D. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Nature, 504, 2013
|
|
1BZH
 
 | | Cyclic peptide inhibitor of human PTP1B | | Descriptor: | PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE 1B INHIBITOR), PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE 1B) | | Authors: | Groves, M.R, Yao, Z.J, Burke Jr, T.R, Barford, D. | | Deposit date: | 1998-10-28 | | Release date: | 1999-02-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
1BZC
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH TPI | | Descriptor: | 4-CARBAMOYL-4-{[6-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALENE-2-CARBONYL]-AMINO}-BUTYRIC ACID, PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE) | | Authors: | Groves, M.R, Yao, Z.J, Jr Burke, T.R, Barford, D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
1A17
 
 | | TETRATRICOPEPTIDE REPEATS OF PROTEIN PHOSPHATASE 5 | | Descriptor: | SERINE/THREONINE PROTEIN PHOSPHATASE 5, SULFATE ION | | Authors: | Das, A.K, Cohen, P.T.W, Barford, D. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions.
EMBO J., 17, 1998
|
|
