8UUW
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12145 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-{(1R)-1-[(3P,5M)-3-(1-methyl-1H-pyrazol-4-yl)-5-(1,3-thiazol-5-yl)phenyl]ethyl}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUF
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUU
 
 | | SARS-Cov-2 papain-like protease (PLpro) with inhibitor Jun12162 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UVM
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with covalent inhibitor Jun11313 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Ansari, A, Tan, B, Chopra, A, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUV
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12197 | | Descriptor: | CHLORIDE ION, N-{(1R)-1-[(3P,5P)-3-(1-cyclopropyl-1H-pyrazol-4-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, Papain-like protease nsp3, ... | | Authors: | Ansari, A, Tan, B, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
1KXG
 
 | | The 2.0 Ang Resolution Structure of BLyS, B Lymphocyte Stimulator. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, B lymphocyte stimulator, CITRIC ACID, ... | | Authors: | Oren, D.A, Li, Y, Volovik, Y, Morris, T.S, Dharia, C, Das, K, Galperina, O, Gentz, R, Arnold, E. | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of BLyS receptor recognition.
Nat.Struct.Biol., 9, 2002
|
|
3KLF
 
 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CYM
 
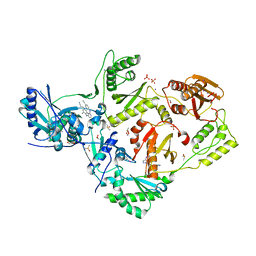 | | HIV-1 reverse transcriptase complexed with 4-iodopyrazole | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
1R0A
 
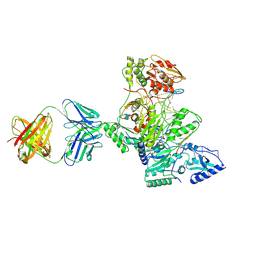 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | Descriptor: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | Authors: | Tuske, S, Ding, J, Arnold, E. | | Deposit date: | 2003-09-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
3QO9
 
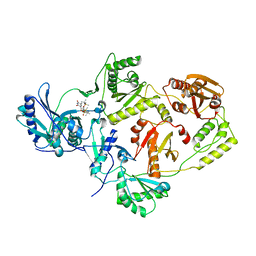 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with TSAO-T, a non-nucleoside RT inhibitor (NNRTI) | | Descriptor: | 1-[(5R,6R,8R,9R)-4-amino-9-{[tert-butyl(dimethyl)silyl]oxy}-6-({[tert-butyl(dimethyl)silyl]oxy}methyl)-2,2-dioxido-1,7-dioxa-2-thiaspiro[4.4]non-3-en-8-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, Reverse transcriptase/ ribonuclease H, p51 RT | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of tert-Butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in Complex with HIV-1 Reverse Transcriptase (RT) Redefines the Elastic Limits of the Non-nucleoside Inhibitor-Binding Pocket.
J.Med.Chem., 54, 2011
|
|
5CXR
 
 | |
1NFG
 
 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
1P91
 
 | | Crystal Structure of RlmA(I) enzyme: 23S rRNA n1-G745 methyltransferase (NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER19) | | Descriptor: | Ribosomal RNA large subunit methyltransferase A, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Das, K, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1N5Y
 
 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
3K2P
 
 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
6ELI
 
 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
1N6Q
 
 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1IHN
 
 | |
3KLG
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to pre-translocation AZTMP-Terminated DNA (COMPLEX N) | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), Reverse transcriptase/ribonuclease H, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1LXY
 
 | | Crystal Structure of Arginine Deiminase covalently linked with L-citrulline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine Deiminase, CITRULLINE | | Authors: | Das, K, Buttler, G.H, Kwiatkowski, V, Yadav, P, Arnold, E. | | Deposit date: | 2002-06-06 | | Release date: | 2004-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism
Structure, 12, 2004
|
|
4IFY
 
 | | HIV-1 reverse transcriptase with bound fragment at the Knuckles site | | Descriptor: | 1-[4-(trifluoromethoxy)phenyl]methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IG3
 
 | | HIV-1 reverse transcriptase with bound fragment near Knuckles site | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
