5BPZ
 
 | | Atomic-resolution structures of the APC/C subunits Apc4 and the Apc5 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Anapc5 protein | | Authors: | Cronin, N, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Atomic-Resolution Structures of the APC/C Subunits Apc4 and the Apc5 N-Terminal Domain.
J.Mol.Biol., 427, 2015
|
|
3MGM
 
 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
5BPT
 
 | |
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
7TQA
 
 | | Crystal Structure of monoclonal S9.6 Fab | | Descriptor: | Fab S9.6 heavy chain, Fab S9.6 light chain, GLYCEROL, ... | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis of R-loop recognition by the S9.6 monoclonal antibody.
Nat Commun, 13, 2022
|
|
6GYU
 
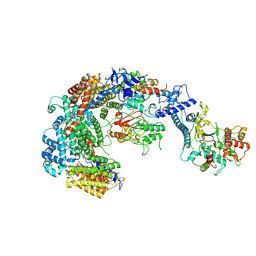 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7TQL
 
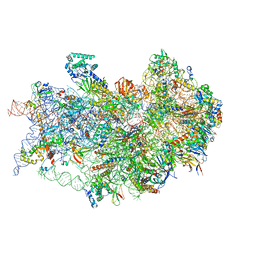 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
4GU0
 
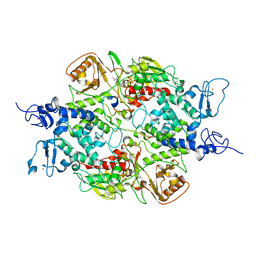 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
6JCJ
 
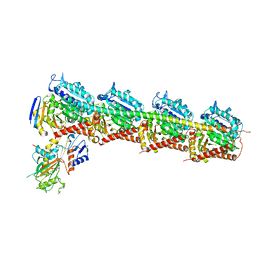 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7XIV
 
 | | Structural insight into the interactions between Lloviu virus VP30 and nucleoprotein | | Descriptor: | Nucleocapsid protein,Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Qin, X.C, Sun, W.Y, Luan, F.C, Wang, J.J, Ma, L, Li, X.X, Yang, G.X, Hao, C.Y. | | Deposit date: | 2022-04-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural insights into the interactions between lloviu virus VP30 and nucleoprotein.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
4LXV
 
 | | Crystal Structure of the Hemagglutinin from a H1N1pdm A/WASHINGTON/5/2011 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Chang, J.C, Guo, Z, Carney, P.J, Shore, D.A, Donis, R.O, Cox, N.J, Villanueva, J.M, Klimov, A.I, Stevens, J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-02-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
J.Virol., 88, 2014
|
|
3F78
 
 | | Crystal structure of wild type LFA1 I domain complexed with isoflurane | | Descriptor: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, GLYCEROL, Integrin alpha-L, ... | | Authors: | Zhang, H, Wang, J.-H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoflurane bound to integrin LFA-1 supports a unified mechanism of volatile anesthetic action in the immune and central nervous systems.
Faseb J., 23, 2009
|
|
3F74
 
 | | Crystal structure of wild type LFA1 I domain | | Descriptor: | GLYCEROL, Integrin alpha-L, MAGNESIUM ION | | Authors: | Zhang, H, Wang, J.-H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of isoflurane bound to integrin LFA-1 supports a unified mechanism of volatile anesthetic action in the immune and central nervous systems.
Faseb J., 23, 2009
|
|
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
4DDP
 
 | |
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
2Q06
 
 | | Crystal structure of Influenza A Virus H5N1 Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Tan, K, Wang, J, Shaw, P.C. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design.
Faseb J., 22, 2008
|
|
2LRW
 
 | | Solution structure of a ubiquitin-like protein from Trypanosoma brucei | | Descriptor: | Ubiquitin, putative | | Authors: | Wang, R, Wang, T, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a ubiqutin-like protein from Trypanosoma bucei
To be Published
|
|
8IQX
 
 | | ferritin mutant-P156H | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
6UP2
 
 | |
4M9Y
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
5XI7
 
 | | Crystal structure of T2R-TTL bound with PO-7 | | Descriptor: | (6Z)-3-[[2,5-bis(fluoranyl)phenyl]methylidene]-6-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Chu, Y, Wang, Y, Yang, J, Li, W. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
5XKF
 
 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
