5JEE
 
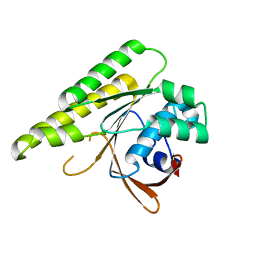 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J2R
 
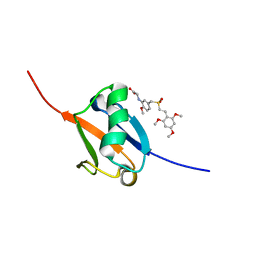 | | Solution structure of Ras Binding Domain (RBD) of B-Raf | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
5MY0
 
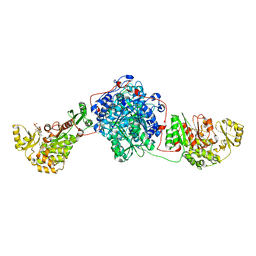 | | KS-MAT DI-DOMAIN OF MOUSE FAS WITH MALONYL-COA | | Descriptor: | COENZYME A, Fatty acid synthase, MALONYL-COENZYME A | | Authors: | Paithankar, K.S, Rittner, A, Huu, K.V, Grininger, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the Polyspecific Transferase of Murine Type I Fatty Acid Synthase (FAS) and Implications for Polyketide Synthase (PKS) Engineering.
ACS Chem. Biol., 13, 2018
|
|
6MG5
 
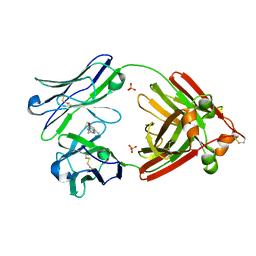 | | Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, Light chain JTO, PHOSPHATE ION | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3NF1
 
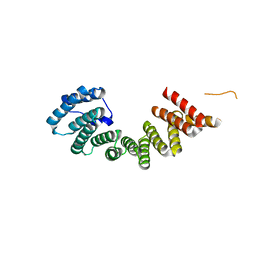 | | Crystal structure of the TPR domain of kinesin light chain 1 | | Descriptor: | Kinesin light chain 1 | | Authors: | Tong, Y, Tempel, W, Shen, L, Shen, Y, Nedyalkova, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-09 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the TPR domain of kinesin light chain 1
to be published
|
|
6MBD
 
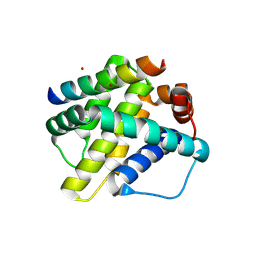 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6FDE
 
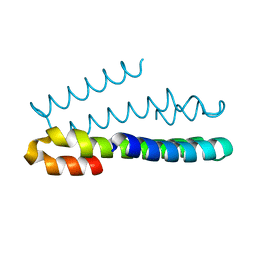 | | Crystal Structure of the HHD2 Domain of Whirlin : 3-helix conformation | | Descriptor: | Whirlin | | Authors: | Delhommel, F, Cordier, F, Saul, F, Haouz, A, Wolff, N. | | Deposit date: | 2017-12-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural plasticity of the HHD2 domain of whirlin.
FEBS J., 285, 2018
|
|
5J51
 
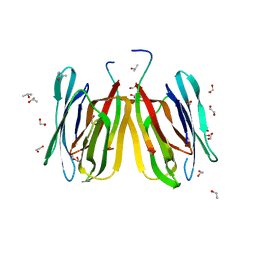 | | Structure of tetrameric jacalin complexed with Gal alpha-(1,4) Gal | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of linkage on the location of reducing and nonreducing sugars bound to jacalin.
IUBMB Life, 68, 2016
|
|
7K9T
 
 | | Co-crystal structure of alpha glucosidase with compound 5 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
4WOD
 
 | | The duplicated taurocyamine kinase from Schistosoma mansoni complexed with arginine | | Descriptor: | ARGININE, Taurocyamine kinase | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
8BOT
 
 | |
6UZN
 
 | |
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
7K9Q
 
 | | Co-crystal structure of alpha glucosidase with compound 4 | | Descriptor: | (1S,2S,3R,4S,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
5T1I
 
 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7KAD
 
 | | Co-crystal structure of alpha glucosidase with compound 6 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
5JGA
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 11c | | Descriptor: | N-[5-(4-methylpiperazine-1-carbonyl)[1,1'-biphenyl]-2-yl]-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
7JTY
 
 | | Co-crystal structure of alpha glucosidase with compound 1 | | Descriptor: | (1S,2S,3R,4S,5S)-5-(butylamino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
5FWS
 
 | | Wnt modulator Kremen crystal form I at 1.90A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
7MTU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P221 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P221
To Be Published
|
|
7JTO
 
 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
8DY7
 
 | |
6ZOY
 
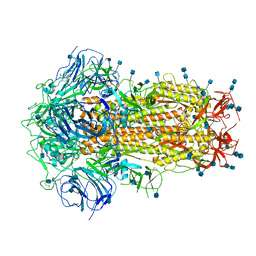 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I5W
 
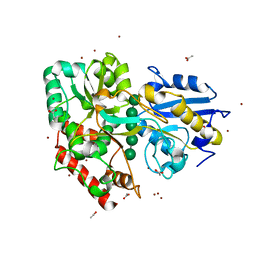 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | ACETATE ION, Solute Binding Protein BlMnBP1, Blac_00780 in complex with mannopentaose, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
