4YBT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | Descriptor: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAX
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | Descriptor: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4MUW
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4MVH
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 1H-benzimidazol-2-yl(4-{[3-(morpholin-4-yl)pyrazin-2-yl]oxy}phenyl)methanone, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-23 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4ZHJ
 
 | |
4IZG
 
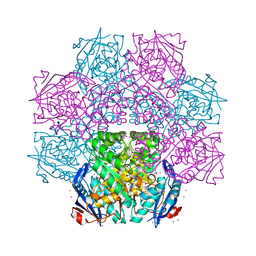 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | Descriptor: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
6BTN
 
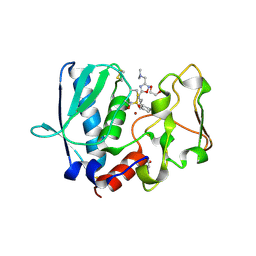 | | BMP1 complexed with a reverse hydroxymate - compound 1 | | Descriptor: | (3S)-6-cyclohexyl-3-{3-[(dimethylamino)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyhexanamide, 1,2-ETHANEDIOL, Bone morphogenetic protein 1, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BSM
 
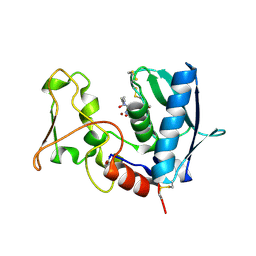 | | BMP1 complexed with a reverse hydroxamate - compound 4 | | Descriptor: | Bone morphogenetic protein 1, N-({[(2R)-2-{[hydroxy(hydroxymethyl)amino]methyl}heptanoyl]amino}methyl)-7-methoxy-1-benzofuran-2-carboxamide, ZINC ION | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTO
 
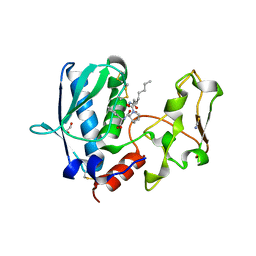 | | BMP1 complexed with (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid | | Descriptor: | (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid, 1,2-ETHANEDIOL, Bone morphogenetic protein 1, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTP
 
 | | BMP1 complexed with a hydroxamate | | Descriptor: | (1R,3S,4S)-2-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]heptanoyl}-N-(3-methoxypyrazin-2-yl)-2-azabicyclo[2.2.1]heptane-3-carboxamide, Bone morphogenetic protein 1, THIOCYANATE ION, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
4J1O
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
6BSL
 
 | | BMP1 complexed with a reverse hydroxymate - compound 22 | | Descriptor: | 1,2-ETHANEDIOL, Bone morphogenetic protein 1, N-(2-ethoxy-4-{5-[({[(2R)-2-{(1R)-1-[formyl(hydroxy)amino]propyl}heptanoyl]amino}methyl)carbamoyl]furan-2-yl}benzene-1-carbonyl)-L-aspartic acid, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTQ
 
 | | BMP1 complexed with a hydroxamate - compound 2 | | Descriptor: | 1,2-ETHANEDIOL, Bone morphogenetic protein 1, N~2~-[(2H-1,3-benzodioxol-5-yl)methyl]-N-hydroxy-N~2~-[(4-methoxyphenyl)sulfonyl]-3-thiophen-2-yl-D-alaninamide, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
4PS4
 
 | | Crystal structure of the complex between IL-13 and M1295 FAB | | Descriptor: | Interleukin-13, M1295 HEAVY CHAIN, M1295 LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Framework Adaptation of a Mouse Anti-Human Il-13 Antibody.
J.Mol.Biol., 398, 2010
|
|
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5GND
 
 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
5GXU
 
 | | Cystal structure of Arabidopsis ATR2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH--cytochrome P450 reductase 2 | | Authors: | Niu, G, Liu, L. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function
FEBS J., 284, 2017
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | Descriptor: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6K
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638, Target EFI-510113 (Desal_0342), complex with diglycerolphosphate | | Descriptor: | TRAP dicarboxylate transporter-DctP subunit, bis[(2S)-2,3-dihydroxypropyl] hydrogen phosphate | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-13 | | Release date: | 2013-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NGU
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio alaskensis G20 (Dde_1548), Target EFI-510103, with bound D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-02 | | Release date: | 2013-12-04 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
