6XH7
 
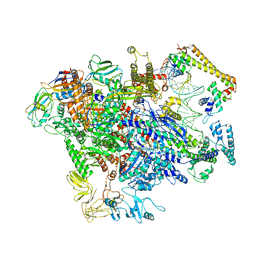 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
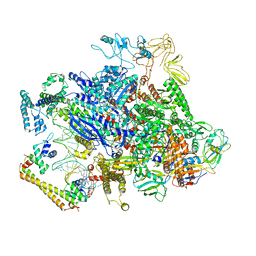 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6PMJ
 
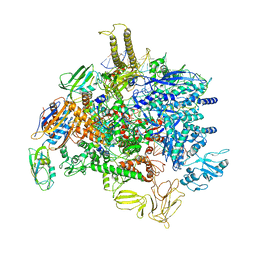 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6PMI
 
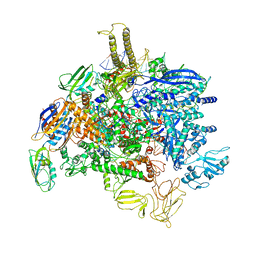 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
6PB4
 
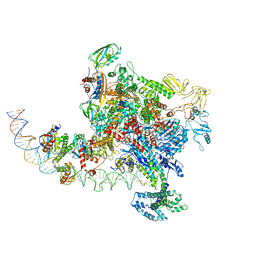 | |
6PB5
 
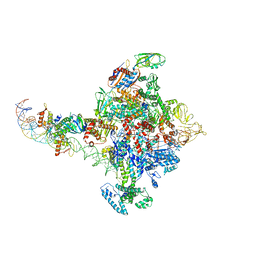 | |
6PB6
 
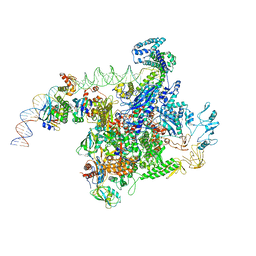 | |
4P9U
 
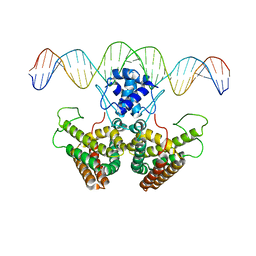 | |
1K27
 
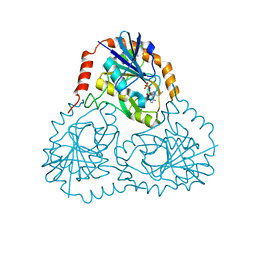 | | Crystal Structure of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase in Complex with a Transition State Analogue | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-Deoxy-5'-Methylthioadenosine Phosphorylase, PHOSPHATE ION | | Authors: | Shi, W, Singh, V, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-09-26 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Picomolar transition state analogue inhibitors of human 5'-methylthioadenosine phosphorylase and X-ray structure with MT-immucillin-A
Biochemistry, 43, 2004
|
|
1ZOS
 
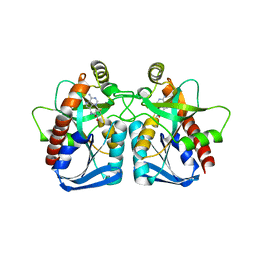 | | Structure of 5'-methylthionadenosine/S-Adenosylhomocysteine nucleosidase from S. pneumoniae with a transition-state inhibitor MT-ImmA | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase | | Authors: | Shi, W, Singh, V, Zhen, R, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2005-05-13 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae.
Biochemistry, 45, 2006
|
|
7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
7DH5
 
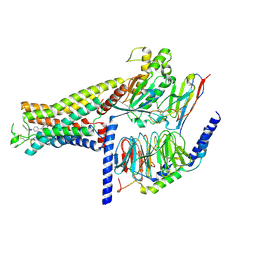 | | Dog beta3 adrenergic receptor bound to mirabegron in complex with a miniGs heterotrimer | | Descriptor: | 2-(2-azanyl-1,3-thiazol-4-yl)-N-[4-[2-[[(2R)-2-oxidanyl-2-phenyl-ethyl]amino]ethyl]phenyl]ethanamide, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
4MMH
 
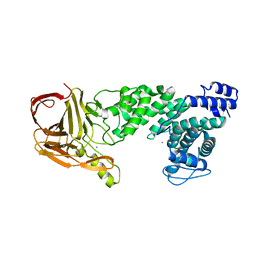 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MMI
 
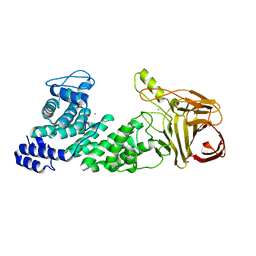 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
7XJI
 
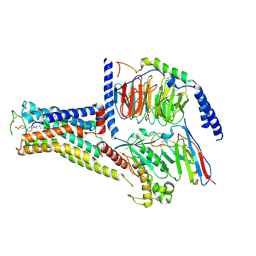 | | Solabegron-activated dog beta3 adrenergic receptor | | Descriptor: | 3-[3-[2-[[(2~{S})-2-(3-chlorophenyl)-2-oxidanyl-ethyl]amino]ethylamino]phenyl]benzoic acid, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the beta 3 adrenergic receptor bound to solabegron and isoproterenol.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
7E4G
 
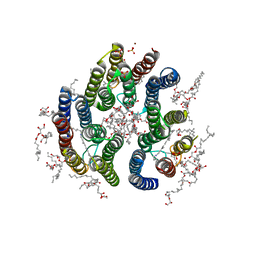 | | Crystal structure of schizorhodopsin 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, SULFATE ION, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2021-02-12 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of schizorhodopsin reveals mechanism of inward proton pumping.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FGT
 
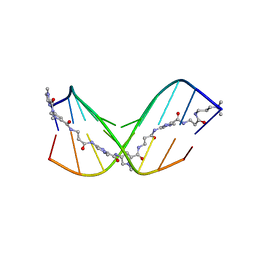 | | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA) | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*CP*TP*T)-3'), ~{N}-[3-[3-(dimethylamino)propylamino]-3-oxidanylidene-propyl]-1-methyl-4-[3-[[1-methyl-4-[[1-methyl-4-[3-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazole-2-carboxamide | | Authors: | Takase, N, Kawai, G, Igarashi, W, Katagiri, T. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA)
To be published
|
|
5Y9J
 
 | | BAFF in complex with belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belibumab light chain, belimumab heavy chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
5Y9K
 
 | | Structure of the belimumab Fab fragment | | Descriptor: | belimumab heavy chain, belimumab light chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
2ZYC
 
 | | Crystal structure of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | PHOSPHATE ION, Peptidoglycan hydrolase FlgJ | | Authors: | Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3AY2
 
 | | Crystal structure of Neisserial azurin | | Descriptor: | GLYCEROL, Lipid modified azurin protein, SULFATE ION, ... | | Authors: | Ochiai, A, Hashimoto, W, Yamada, T, Chakrabarty, A.M, Murata, K. | | Deposit date: | 2011-04-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Neisserial Azurin
To be Published
|
|
4O59
 
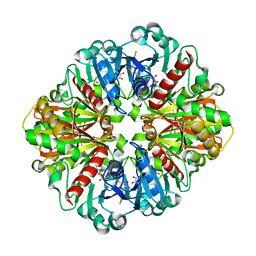 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4O63
 
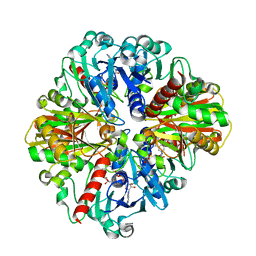 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
