6PX8
 
 | |
5G4C
 
 | | Human SIRT2 catalyse short chain fatty acyl lysine | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, SIRT2, ... | | Authors: | Wang, Y. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones.
J. Am. Chem. Soc., 138, 2016
|
|
8HN9
 
 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
7T7M
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7L
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
4X3O
 
 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Wang, Y, Zhang, W.Z, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
4X3P
 
 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, Y, Zhang, W, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
8C13
 
 | | Crystal structure of pVHL:ElonginC:ElonginB complex bound to PROTAC JW48 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[3-[2-[2-[2-(acetamidomethyl)-4-(6,7-dihydro-5~{H}-pyrrolo[1,2-a]imidazol-2-yl)phenoxy]ethoxy]ethoxy]propanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Weckesser, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
6P7I
 
 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
7S8L
 
 | | CryoEM structure of Gq-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8N
 
 | | CryoEM structure of Gq-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8P
 
 | | CryoEM structure of Gq-coupled MRGPRX4 with small molecule agonist MS47134 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8M
 
 | | CryoEM structure of Gi-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8O
 
 | | CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7Q2J
 
 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
6R2S
 
 | |
6RCV
 
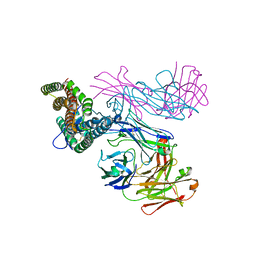 | | PfRH5 bound to monoclonal antibodies R5.011 and R5.016 | | Descriptor: | R5.011 heavy chain, R5.011 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCO
 
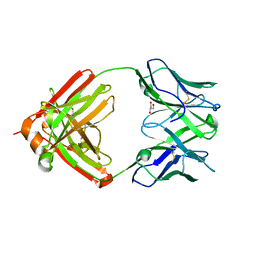 | | PfRH5-binding monoclonal antibody R5.004 | | Descriptor: | ACETATE ION, GLYCEROL, R5.004 heavy chain, ... | | Authors: | Alanine, D.G.W, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCU
 
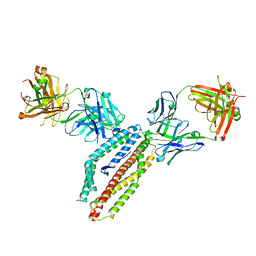 | | PfRH5 bound to monoclonal antibodies R5.004 and R5.016 | | Descriptor: | R5.004 heavy chain, R5.004 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCQ
 
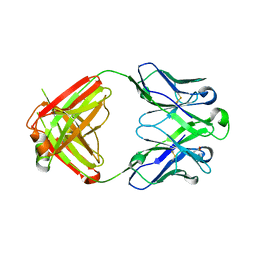 | | PfRH5-binding monoclonal antibody R5.011 | | Descriptor: | ACETATE ION, CACODYLATE ION, R5.011 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCS
 
 | | PfRH5-binding monoclonal antibody R5.016 | | Descriptor: | ACETATE ION, R5.016 heavy chain, R5.016 light chain, ... | | Authors: | Alanine, D.W.G, Jamwal, A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
4R0T
 
 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | Descriptor: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
4R0S
 
 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
3FDR
 
 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
