1Y8X
 
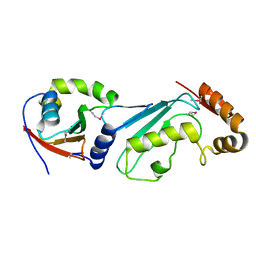 | | Structural basis for recruitment of Ubc12 by an E2-binding domain in NEDD8's E1 | | Descriptor: | Ubiquitin-activating enzyme E1C, Ubiquitin-conjugating enzyme E2 M | | Authors: | Huang, D.T, Paydar, A, Zhuang, M, Waddell, M.B, Holton, J.M, Schulman, B.A. | | Deposit date: | 2004-12-13 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
Mol.Cell, 17, 2005
|
|
2NVU
 
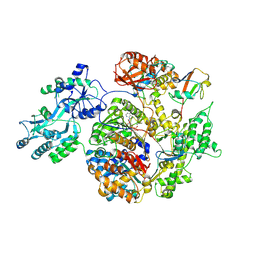 | | Structure of APPBP1-UBA3~NEDD8-NEDD8-MgATP-Ubc12(C111A), a trapped ubiquitin-like protein activation complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera, ... | | Authors: | Huang, D.T, Hunt, H.W, Zhuang, M, Ohi, M.D, Holton, J.M, Schulman, B.A. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Nature, 445, 2007
|
|
3FN1
 
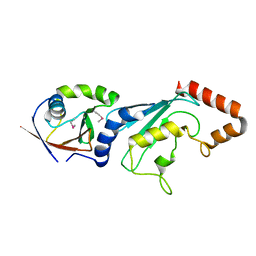 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
1TT5
 
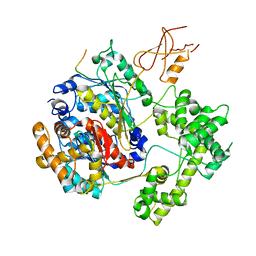 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5J3X
 
 | | Structure of c-CBL Y371F | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, D.T, Buetow, L, Dou, H. | | Deposit date: | 2016-03-31 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation.
Bmc Biol., 14, 2016
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1K8O
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
4V3K
 
 | | RNF38-UbcH5B-UB complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF38, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
4V3L
 
 | | RNF38-UB-UbcH5B-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF38, POLYUBIQUITIN-C, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
5O76
 
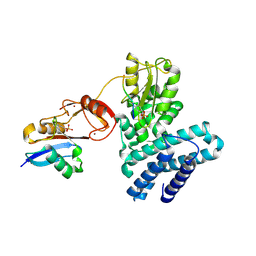 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5O6T
 
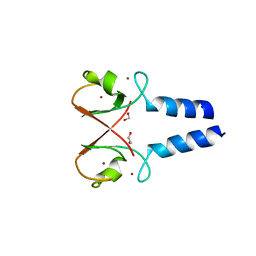 | | BIRC4 RING in complex with dimeric ubiquitin variant | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase XIAP, Polyubiquitin-B, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5O6S
 
 | |
5O75
 
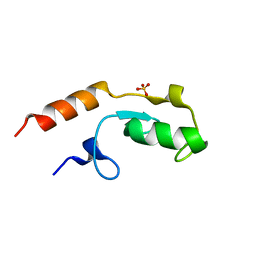 | | Ube4B U-box domain | | Descriptor: | SULFATE ION, Ubiquitin conjugation factor E4 B | | Authors: | Gabrielsen, M, Buetow, L, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
6HPR
 
 | |
5MNJ
 
 | | Structure of MDM2-MDMX-UbcH5B-ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Polyubiquitin-B, Protein Mdm4, ... | | Authors: | Klejnot, M, Huang, D.T. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of MDM2 RING separates degradation from regulation of p53 transcription activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
9FJ3
 
 | | Structure of ubiquitin bound of coiled-coil UIM form 2 | | Descriptor: | 3-[1-(2-oxidanylideneethyl)-1,2,3-triazol-4-yl]propanal, GLU-GLN-GLU-ILE-GLU-GLU-LEU-GLU-ILE-GLU-ILE-ALA-ILE-LEU-LEU-SER-GLU-ILE-GLU-GLY, LYS-GLN-LYS-ILE-ALA-ALA-LEU-LYS-TYR-LYS-ILE-ALA-ALA-LEU-LYS-GLN-LYS-ILE, ... | | Authors: | Paredes Vergara, P, Huang, D.T. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An engineered ubiquitin binding coiled coil peptide.
Chem Sci, 15, 2024
|
|
9FJ4
 
 | | Structure of ubiquitin bound to coiled coil-UIM form 1 | | Descriptor: | 3-[1-(2-oxidanylideneethyl)-1,2,3-triazol-4-yl]propanal, GLU-GLN-GLU-ILE-GLU-GLU-LEU-GLU-ILE-GLU-ILE-ALA-ILE-LEU-LEU-SER-GLU-ILE-GLU-GLY, LYS-GLN-LYS-ILE-ALA-ALA-LEU-LYS-TYR-LYS-ILE-ALA-ALA-LEU-LYS-GLN-LYS-ILE-GLN, ... | | Authors: | Paredes Vergara, P, Huang, D.T. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | An engineered ubiquitin binding coiled coil peptide.
Chem Sci, 15, 2024
|
|
1ZWV
 
 | |
3RNM
 
 | | The crystal structure of the subunit binding of human dihydrolipoamide transacylase (E2b) bound to human dihydrolipoamide dehydrogenase (E3) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-MERCAPTOETHANOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Brautigam, C.A, Wynn, R.M, Chuang, J.C, Young, B.B, Chuang, D.T. | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Basis for Weak Interactions between Dihydrolipoamide Dehydrogenase and Subunit-binding Domain of the Branched-chain {alpha}-Ketoacid Dehydrogenase Complex.
J.Biol.Chem., 286, 2011
|
|
6QK9
 
 | |
7OJX
 
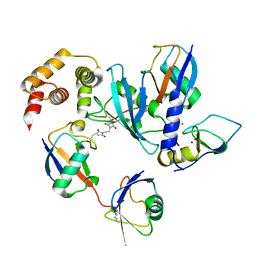 | | E2 UBE2K covalently linked to donor Ub, acceptor di-Ub, and RING E3 primed for K48-linked Ub chain synthesis | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), E3 ubiquitin-protein ligase RNF38, Polyubiquitin-B, ... | | Authors: | Majorek, K.A, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of UBE2K-Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension.
Nat.Chem.Biol., 18, 2022
|
|
6Y2X
 
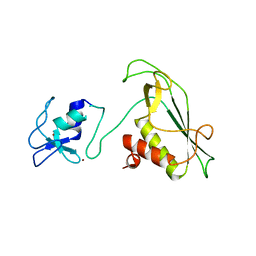 | | RING-DTC domains of Deltex 2, Form 2 | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
6Y3J
 
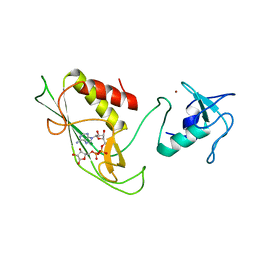 | | RING-DTC domains of Deltex 2, bound to ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielssen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
6Y5P
 
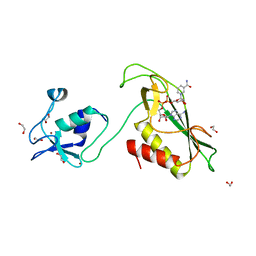 | | RING-DTC domain of Deltex1 bound to NAD | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase DTX1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
2Y1M
 
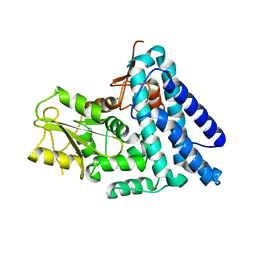 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
