9R2O
 
 | | De novo designed N5 protein fold | | Descriptor: | N5, POTASSIUM ION, THIOCYANATE ION | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2L
 
 | | De novo designed N37 protein fold | | Descriptor: | ACETATE ION, ETHANOLAMINE, N37, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2V
 
 | | De novo designed M16 protein fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-05-01 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2R
 
 | |
9R2K
 
 | |
8PI3
 
 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
7XAD
 
 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8OYS
 
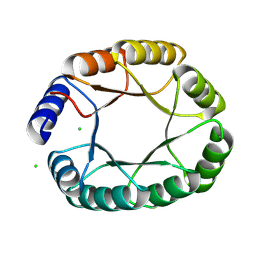 | | De novo designed TIM barrel fold TBF_24 | | Descriptor: | CHLORIDE ION, De novo designed TIM-barrel | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYV
 
 | | De novo designed Claudin fold CLF_4 | | Descriptor: | De novo designed soluble Claudin | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYY
 
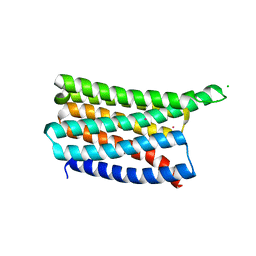 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYW
 
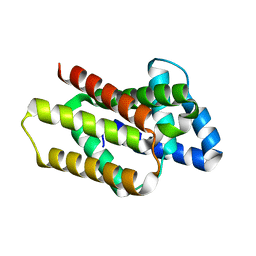 | |
9BEI
 
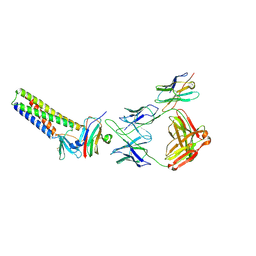 | |
6S3D
 
 | |
7AYE
 
 | | Crystal structure of the computationally designed chemically disruptable heterodimer LD6-MDM2 | | Descriptor: | Isoform 11 of E3 ubiquitin-protein ligase Mdm2, Thiol:disulfide interchange protein DsbD | | Authors: | Yang, C, Lau, K, Pojer, F, Correia, B.E. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A rational blueprint for the design of chemically-controlled protein switches.
Nat Commun, 12, 2021
|
|
6HTF
 
 | |
6VTW
 
 | |
6XWI
 
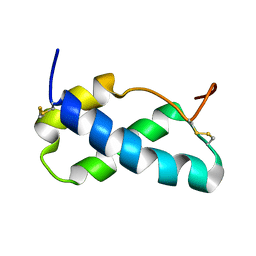 | |
9MER
 
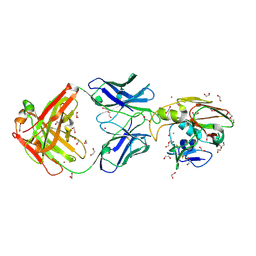 | | Structure of H1H5:FluA20 Chimeric Influenza HA Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Seraj, N. | | Deposit date: | 2024-12-08 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based Design of Chimeric Influenza Hemagglutinins to Elicit Cross-group Immunity.
Biorxiv, 2025
|
|
9MEV
 
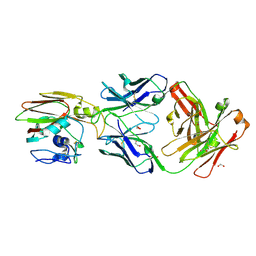 | | Structure of H1H3:FluA20 Chimeric Antigen Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Seraj, N. | | Deposit date: | 2024-12-08 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based Design of Chimeric Influenza Hemagglutinins to Elicit Cross-group Immunity.
Biorxiv, 2025
|
|
6XXV
 
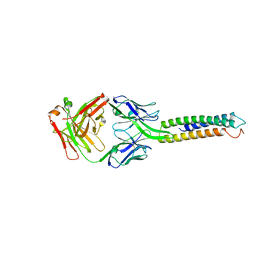 | | Crystal Structure of a computationally designed Immunogen S2_1.2 in complex with its elicited antibody C57 | | Descriptor: | Antibody C57, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Correia, B.E, Pojer, F. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20116425 Å) | | Cite: | De novo protein design enables the precise induction of RSV-neutralizing antibodies.
Science, 368, 2020
|
|
4JEG
 
 | | Crystal Structure of Monobody CS1/SHP2 C-SH2 Domain Complex | | Descriptor: | Monobody CS1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JE4
 
 | | Crystal Structure of Monobody NSa1/SHP2 N-SH2 Domain Complex | | Descriptor: | Monobody NSa1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
