3MXO
 
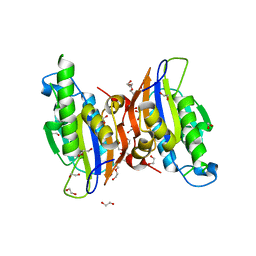 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
2A94
 
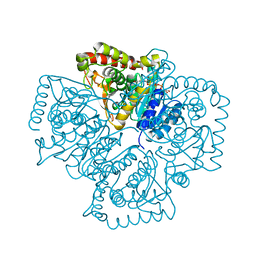 | | Structure of Plasmodium falciparum lactate dehydrogenase complexed to APADH. | | Descriptor: | ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2AA3
 
 | | Crystal structure of Plasmodium vivax lactate dehydrogenase complex with APADH | | Descriptor: | ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, L-lactate dehydrogenase, ... | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2A92
 
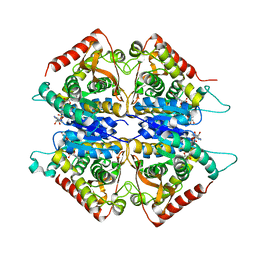 | | Crystal structure of lactate dehydrogenase from Plasmodium vivax: complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
6QAT
 
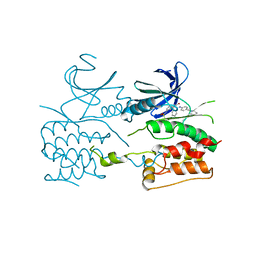 | | Crystal structure of ULK2 in complexed with hesperadin | | Descriptor: | N-{(3Z)-2-oxo-3-[phenyl({4-[(piperidin-1-yl)methyl]phenyl}amino)methylidene]-2,3-dihydro-1H-indol-5-yl}ethanesulfonamide, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAS
 
 | | Crystal structure of ULK1 in complexed with PF-03814735 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAU
 
 | | Crystal structure of ULK2 in complexed with MRT67307 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
4DYM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135 | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Activin receptor type-1, GLYCEROL, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Canning, P, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Krojer, T, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135
To be Published
|
|
6QAV
 
 | | Crystal structure of ULK2 in complexed with MRT68921 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
4FR4
 
 | | Crystal structure of human serine/threonine-protein kinase 32A (YANK1) | | Descriptor: | 1,2-ETHANEDIOL, STAUROSPORINE, Serine/threonine-protein kinase 32A | | Authors: | Chaikuad, A, Elkins, J.M, Krojer, T, Mahajan, P, Goubin, S, Szklarz, M, Tumber, A, Wang, J, Savitsky, P, Shrestha, B, Daga, N, Picaud, S, Fedorov, O, Allerston, C.K, Latwiel, S.V.A, Vollmar, M, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human serine/threonine-protein kinase 32A (YANK1)
To be Published
|
|
4C58
 
 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | Descriptor: | 1,2-ETHANEDIOL, 9-HYDROXY-4-PHENYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
4C59
 
 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cyclin-G-associated kinase, NANOBODY | | Authors: | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
4C57
 
 | | Structure of GAK kinase in complex with a nanobody | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, 1,2-ETHANEDIOL, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Keates, T, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Cyclin G-Associated Kinase (Gak) Trapped in Different Conformations Using Nanobodies.
Biochem.J., 459, 2014
|
|
4XWX
 
 | | Crystal structure of the PTB domain of SHC | | Descriptor: | 1,2-ETHANEDIOL, SHC-transforming protein 1, SODIUM ION | | Authors: | Chaikuad, A, Tallant, C, Krojer, T, Dixon-Clarke, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the PTB domain of SHC
To Be Published
|
|
5LXD
 
 | | Crystal structure of DYRK2 in complex with EHT 1610 (compound 2) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
5LXC
 
 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
3MDY
 
 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
4DYO
 
 | | Crystal Structure of Human Aspartyl Aminopeptidase (DNPEP) in complex with Aspartic acid Hydroxamate | | Descriptor: | Aspartyl aminopeptidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Pilka, E, Vollmar, M, Krojer, T, Muniz, J.R.C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family.
Bmc Struct.Biol., 12, 2012
|
|
7OPG
 
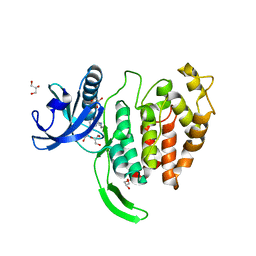 | | Crystal structure of CLK1 in complex with compound 2 (CC513) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[2-(propylamino)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]phenol, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Routier, S, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of CLK1 in complex with compound 2 (CC513)
To Be Published
|
|
6QU2
 
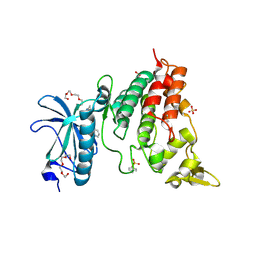 | | Crystal structure of DYRK1A complexed with FC162 inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-cyclopropyl-2-pyridin-3-yl-[1,3]thiazolo[5,4-f]quinazolin-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of DYRK1A complexed with FC162 inhibitor
To Be Published
|
|
6RCG
 
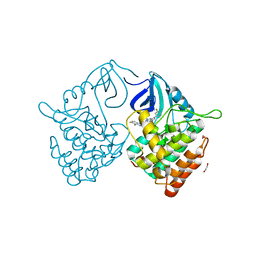 | | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, ~{N}-[[6,7-bis(fluoranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(3-fluorophenyl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor
To Be Published
|
|
6RCH
 
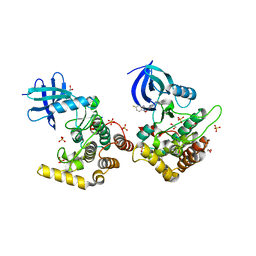 | | Crystal structure of Casein kinase I isoform delta (CK1 delta) complexed with SR4133 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, SODIUM ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of Casein kinase I isoform delta (CK1 delta) complexed with SR4133 inhibitor
To Be Published
|
|
6RCT
 
 | | Crystal structure of CLK3 in complex with T3-CLK | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK3 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of CLK3 in complex with TP003
To Be Published
|
|
3PRY
 
 | | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Chaikuad, A, Pilka, E, Sharpe, T.D, Cooper, C.D.O, Phillips, C, Berridge, G, Ayinampudi, V, Fedorov, O, Keates, T, Thangaratnarajah, C, Zimmermann, T, Vollmar, M, Yue, W.W, Che, K.H, Krojer, T, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution
To be Published
|
|
7QRW
 
 | |
