4IQZ
 
 | | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli | | Descriptor: | DNA-directed RNA polymerase subunit beta', IODIDE ION, SODIUM ION | | Authors: | Bhandari, V, Sugiman-Marangos, S.N, Naushad, H.S, Gupta, R.S, Junop, M.S. | | Deposit date: | 2013-01-14 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli
To be Published
|
|
7UGC
 
 | | Structure of the N-terminal domain of ViaA | | Descriptor: | Protein ViaA | | Authors: | Lemak, A, Reichheld, S, Bhandari, V, Houliston, S, Arrowsmith, C.H, Sharpe, S, Houry, W.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of ViaA
To Be Published
|
|
3KER
 
 | |
8DBB
 
 | | Crystal structure of DDT with the selective inhibitor 2,5-Pyridinedicarboxylic Acid | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase, pyridine-2,5-dicarboxylic acid | | Authors: | Parkins, A, Banumathi, S, Pantouris, G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2,5-Pyridinedicarboxylic acid is a bioactive and highly selective inhibitor of D-dopachrome tautomerase.
Structure, 31, 2023
|
|
4GUM
 
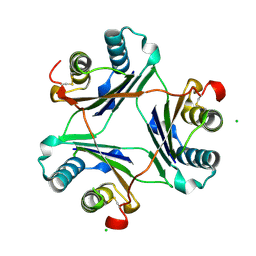 | | Cystal structure of locked-trimer of human MIF | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4TRF
 
 | |
4TRU
 
 | |
4XX7
 
 | |
4XX8
 
 | |
6BG7
 
 | |
6BG6
 
 | |
7KQX
 
 | | MIF Y99C homotrimeric mutant | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Manjula, R, Georgios, P, Lolis, E.J. | | Deposit date: | 2020-11-18 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Cysteine Variant at an Allosteric Site Alters MIF Dynamics and Biological Function in Homo- and Heterotrimeric Assemblies.
Front Mol Biosci, 9, 2022
|
|
5EIZ
 
 | |
5UZY
 
 | |
6NAW
 
 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NB1
 
 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
6NAH
 
 | |
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | Descriptor: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
4P01
 
 | |
4PKZ
 
 | |
