7VDM
 
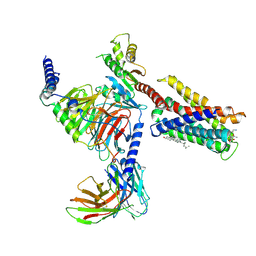 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with substance P | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
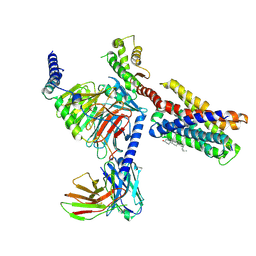
7VDL
 
 | |
7VDH
 
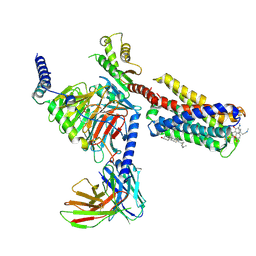 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state2 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV0
 
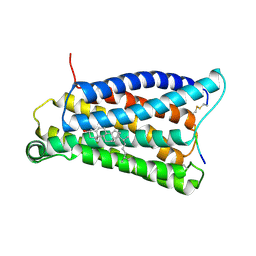 | |
7VV6
 
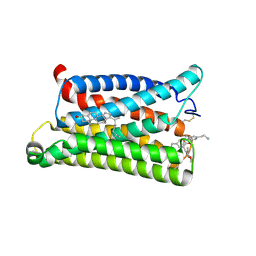 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Mas-related G-protein coupled receptor member X2 | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VUY
 
 | |
7VV3
 
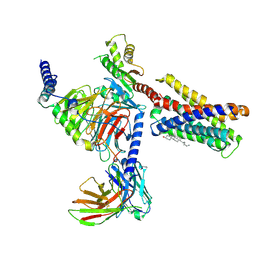 | |
8W8Q
 
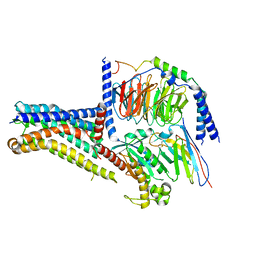 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
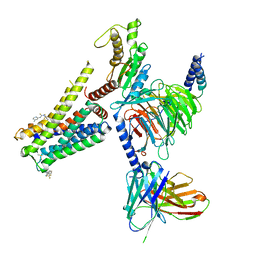 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
1A5W
 
 | | ASV INTEGRASE CORE DOMAIN WITH HIV-1 INTEGRASE INHIBITOR Y3 | | Descriptor: | 4-ACETYLAMINO-5-HYDROXYNAPHTHALENE-2,7-DISULFONIC ACID, INTEGRASE | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of avian sarcoma virus integrase with a bound HIV-1 integrase-targeted inhibitor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A5V
 
 | | ASV INTEGRASE CORE DOMAIN WITH HIV-1 INTEGRASE INHIBITOR Y3 AND MN CATION | | Descriptor: | 4-ACETYLAMINO-5-HYDROXYNAPHTHALENE-2,7-DISULFONIC ACID, INTEGRASE, MANGANESE (II) ION | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the catalytic domain of avian sarcoma virus integrase with a bound HIV-1 integrase-targeted inhibitor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A5X
 
 | | ASV INTEGRASE CORE DOMAIN WITH HIV-1 INTEGRASE INHIBITOR Y3 | | Descriptor: | 4-ACETYLAMINO-5-HYDROXYNAPHTHALENE-2,7-DISULFONIC ACID, INTEGRASE | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the catalytic domain of avian sarcoma virus integrase with a bound HIV-1 integrase-targeted inhibitor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
5Z3R
 
 | | Crystal Structure of Delta 5-3-Ketosteroid Isomerase from Mycobacterium sp. | | Descriptor: | Steroid delta-isomerase | | Authors: | Cheng, X.Y, Peng, F, Yang, F, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-based reconstruction of a Mycobacterium hypothetical protein into an active Delta5-3-ketosteroid isomerase.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
5Z74
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
