2HOS
 
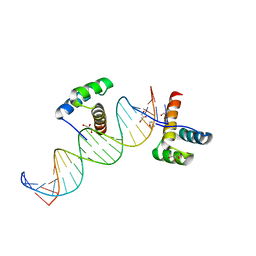 | | Phage-Selected Homeodomain Bound to Unmodified DNA | | Descriptor: | 3-METHYL-1,3-OXAZOLIDIN-2-ONE, 5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*CP*CP*GP*GP*A)-3', ... | | Authors: | Shokat, K.M, Feldman, M.E, Simon, M.D. | | Deposit date: | 2006-07-16 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and properties of a re-engineered homeodomain protein-DNA interface.
Acs Chem.Biol., 1, 2006
|
|
6CZ5
 
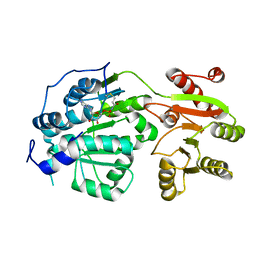 | | Crystal structure of small molecule AMP-acrylamide covalently bound to DDX3 S228C | | Descriptor: | 5'-O-[(R)-hydroxy(propanoylamino)phosphoryl]adenosine, ATP-dependent RNA helicase DDX3X | | Authors: | Barkovich, K.J, Moore, M.K, Hu, Q, Shokat, K.M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical genetic inhibition of DEAD-box proteins using covalent complementarity.
Nucleic Acids Res., 46, 2018
|
|
6P8F
 
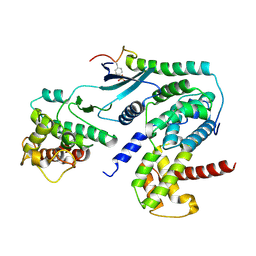 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8H
 
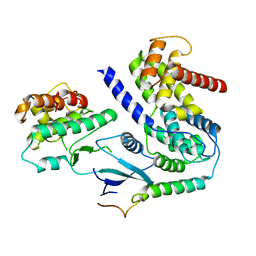 | | Crystal structure of CDK4 in complex with CyclinD1 and P21 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
4WNP
 
 | | Structure of ULK1 bound to a potent inhibitor | | Descriptor: | GLYCEROL, N~2~-(1H-benzimidazol-6-yl)-N~4~-(5-cyclobutyl-1H-pyrazol-3-yl)quinazoline-2,4-diamine, Serine/threonine-protein kinase ULK1 | | Authors: | Lazarus, M.B, Novotny, C.J, Shokat, K.M. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of the Human Autophagy Initiating Kinase ULK1 in Complex with Potent Inhibitors.
Acs Chem.Biol., 10, 2015
|
|
7E5E
 
 | | Crystal structure of GDP-bound GNAS in complex with the cyclic peptide inhibitor GD20 | | Descriptor: | CHLORIDE ION, GD20, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, Q, Dai, S, Shokat, K.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides.
Cell, 185, 2022
|
|
1WMA
 
 | | Crystal structure of human CBR1 in complex with Hydroxy-PP | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, ... | | Authors: | Rauh, D, Bateman, R, Shokat, K.M. | | Deposit date: | 2004-07-06 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An unbiased cell morphology-based screen for new, biologically active small molecules
Plos Biol., 3, 2005
|
|
7BPH
 
 | |
1KSW
 
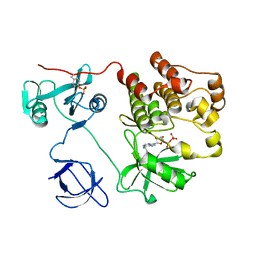 | | Structure of Human c-Src Tyrosine Kinase (Thr338Gly Mutant) in Complex with N6-benzyl ADP | | Descriptor: | N6-BENZYL ADENOSINE-5'-DIPHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Witucki, L.A, Huang, X, Shah, K, Liu, Y, Kyin, S, Eck, M.J, Shokat, K.M. | | Deposit date: | 2002-01-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme.
Chem.Biol., 9, 2002
|
|
6WGN
 
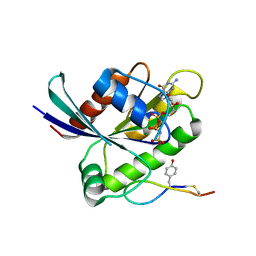 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
8T4V
 
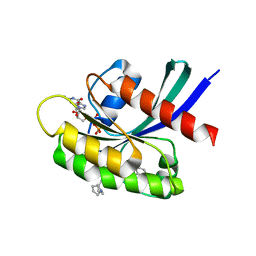 | | Crystal structure of compound 1 bound to K-Ras(G12D) | | Descriptor: | 4-{(1R,5S)-3-[(7P)-7-(8-ethynylnaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl}-4-oxobutanoic acid, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Zheng, Q, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2023-06-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain-release alkylation of Asp12 enables mutant selective targeting of K-Ras-G12D.
Nat.Chem.Biol., 20, 2024
|
|
8TLR
 
 | | Crystal Structure of human HRAS G12C covalently bound to AMG 510 | | Descriptor: | AMG 510 (bound form), GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Morstein, J, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.70003951 Å) | | Cite: | Targeting Ras-, Rho-, and Rab-family GTPases via a conserved cryptic pocket.
Cell, 2024
|
|
2CHX
 
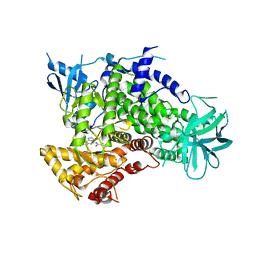 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHZ
 
 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHW
 
 | | A pharmacological map of the PI3-K family defines a role for p110 alpha in signaling: The structure of complex of phosphoinositide 3- kinase gamma with inhibitor PIK-39 | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
4D0L
 
 | | Phosphatidylinositol 4-kinase III beta-PIK93 in a complex with Rab11a- GTP gammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
4D0M
 
 | | Phosphatidylinositol 4-kinase III beta in a complex with Rab11a-GTP- gamma-S and the Rab-binding domain of FIP3 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughlin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
6P8G
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8E
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1, ... | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
8CX5
 
 | | Crystal Structure of small molecule alpha,beta-ketoamide 4 covalently bound to K-Ras(G12R) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Morstein, J, Ecker, A, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Chemoselective Covalent Modification of K-Ras(G12R) with a Small Molecule Electrophile.
J.Am.Chem.Soc., 144, 2022
|
|
5CI7
 
 | | Structure of ULK1 bound to a selective inhibitor | | Descriptor: | GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]-5-iodopyrimidin-2-yl}amino)phenyl]pyrrolidine-1-carboxamide, Serine/threonine-protein kinase ULK1 | | Authors: | Lazarus, M.B, Shokat, K.M. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and structure of a new inhibitor scaffold of the autophagy initiating kinase ULK1.
Bioorg.Med.Chem., 23, 2015
|
|
8DC6
 
 | | Crystal structure of p53 Y220C covalently bound to indole KG6 | | Descriptor: | 1-(2-methylprop-2-enoyl)-1H-indole-3-carbaldehyde, bound form, Cellular tumor antigen p53, ... | | Authors: | Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.60000908 Å) | | Cite: | A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Cancer Discov, 13, 2023
|
|
8DC4
 
 | |
8DC7
 
 | | Crystal structure of p53 Y220C covalently bound to indole KG10 | | Descriptor: | 4-[4-(4-methylpiperazin-1-yl)phenyl]-1-(2-methylprop-2-enoyl)-1H-indole-3-carbaldehyde, bound form, Cellular tumor antigen p53, ... | | Authors: | Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9870069 Å) | | Cite: | A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Cancer Discov, 13, 2023
|
|
8DC8
 
 | | Crystal structure of p53 Y220C covalently bound to azaindole KG13 | | Descriptor: | 2-methyl-1-[(4P)-3-methyl-4-(2-methyl-1,2,3,4-tetrahydroisoquinolin-6-yl)-1H-pyrrolo[2,3-c]pyridin-1-yl]prop-2-en-1-one, bound form, Cellular tumor antigen p53, ... | | Authors: | Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7200973 Å) | | Cite: | A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Cancer Discov, 13, 2023
|
|
