7E20
 
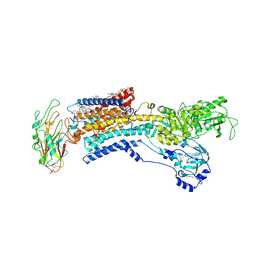 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | 登録日 | 2021-02-04 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7CAJ
 
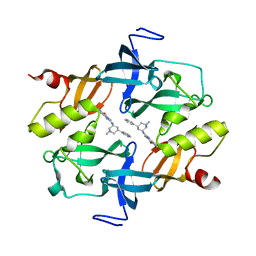 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | 分子名称: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CJT
 
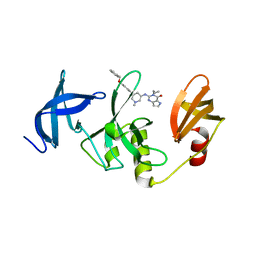 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | 分子名称: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.474 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5JIE
 
 | |
4WD8
 
 | |
4WD7
 
 | |
6OT1
 
 | |
6OSY
 
 | |
7N5H
 
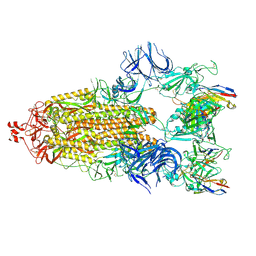 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | 分子名称: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Casner, R.G, Cerutti, G, Shapiro, L. | | 登録日 | 2021-06-05 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7YW0
 
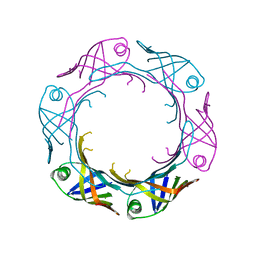 | | Bacteroides fragilis Hcp5 | | 分子名称: | Bacterodales T6SS protein TssD (Hcp) | | 著者 | Wen, Y, He, W, Bai, Y. | | 登録日 | 2022-08-20 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
6PZV
 
 | |
7LSS
 
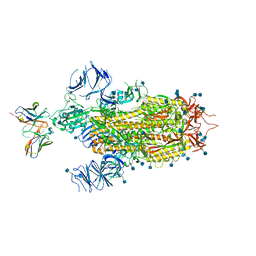 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-7 variable heavy chain, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2021-02-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.
Structure, 29, 2021
|
|
7LS9
 
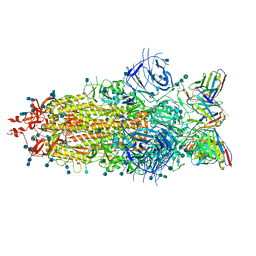 | |
8WNT
 
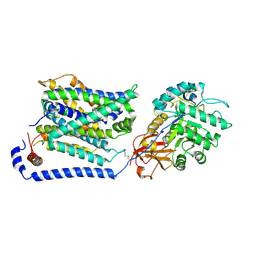 | | Cryo EM map of SLC7A10 with L-Alanine substrate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | 著者 | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | 登録日 | 2023-10-06 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
7L5B
 
 | |
7RW2
 
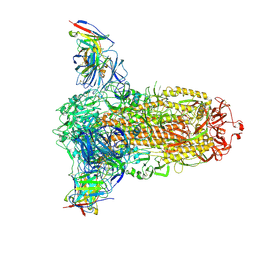 | |
5KF4
 
 | |
8WNS
 
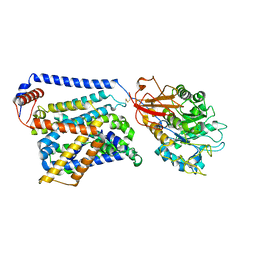 | | Cryo EM map of SLC7A10 in the apo state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A2, Asc-type amino acid transporter 1 | | 著者 | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | 登録日 | 2023-10-06 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
8WNY
 
 | | Cryo EM map of SLC7A10-SLC3A2 complex in the D-serine bound state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A2, Asc-type amino acid transporter 1, ... | | 著者 | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | 登録日 | 2023-10-06 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
6UBI
 
 | |
6UCE
 
 | |
6UCF
 
 | |
6VIL
 
 | |
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | 分子名称: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | 著者 | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-18 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | 著者 | Yan, W, Zhang, Y. | | 登録日 | 2015-02-12 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
