6U2P
 
 | | PCSK9 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
7UM7
 
 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | | Descriptor: | (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM5
 
 | | CryoEM structure of Go-coupled 5-HT5AR in complex with 5-CT | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM4
 
 | | Crystal structure of inactive 5-HT5AR in complex with AS2674723 | | Descriptor: | 5-hydroxytryptamine receptor 5A, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Zhang, S, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM6
 
 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | | Descriptor: | 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4WF6
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor MK-31 | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6521 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
8VY7
 
 | | CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kim, Y, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2024-02-07 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
8VY9
 
 | | CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kim, Y, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2024-02-07 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
4FFW
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4FFV
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
1JL4
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6LOJ
 
 | | The complex structure of IpaH9.8-LRR and hGBP1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 1, Invasion plasmid antigen | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
6LOL
 
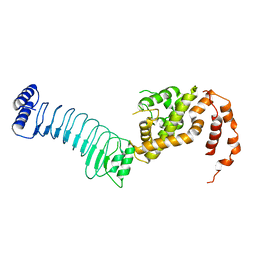 | | The crystal structure of full length IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
7LTT
 
 | | SAMHD1(113-626) H206R D207N R366C | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
7LU5
 
 | | SAMHD1(113-626) H206R D207N R366H | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
6V40
 
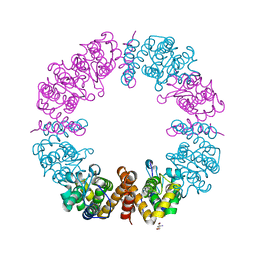 | | Structure of Salmonella Typhi TtsA | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PG_binding_3 domain-containing protein | | Authors: | Galan, J.E, Lara-Tejero, M. | | Deposit date: | 2019-11-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Mechanisms of substrate recognition by a typhoid toxin secretion-associated muramidase.
Elife, 9, 2020
|
|
6WJ6
 
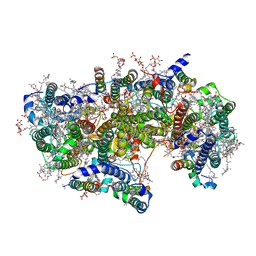 | | Cryo-EM structure of apo-Photosystem II from Synechocystis sp. PCC 6803 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gisriel, C.J. | | Deposit date: | 2020-04-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM Structure of Monomeric Photosystem II from Synechocystis sp. PCC 6803 Lacking the Water-Oxidation Complex
Joule, 2020
|
|
7T7L
 
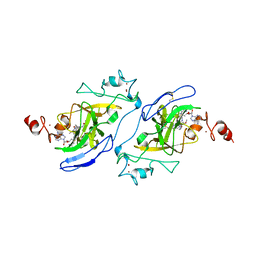 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7M
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
6BWY
 
 | | DNA substrate selection by APOBEC3G | | Descriptor: | DNA (30-MER), PHOSPHATE ION, Protection of telomeres protein 1, ... | | Authors: | Ziegler, S.J, Buzovetsky, O. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into DNA substrate selection by APOBEC3G from structural, biochemical, and functional studies.
PLoS ONE, 13, 2018
|
|
5XDZ
 
 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
5V9O
 
 | | KRAS G12C inhibitor | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C.
Cell Chem Biol, 24, 2017
|
|
5V9L
 
 | |
5Y5W
 
 | |
4HJR
 
 | | Crystal structure of F2YRS | | Descriptor: | Tyrosine-tRNA ligase | | Authors: | Wang, J, Tian, C, Gong, W, Li, F, Shi, P, Li, J, Ding, W. | | Deposit date: | 2012-10-13 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A genetically encoded 19F NMR probe for tyrosine phosphorylation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
