8BAL
 
 | |
8BDP
 
 | |
8BBL
 
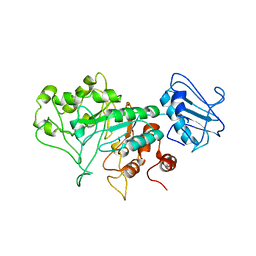 | | SGL a GH20 family sulfoglycosidase | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Dong, M.D, Roth, C.R, Jin, Y.J. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
7NS6
 
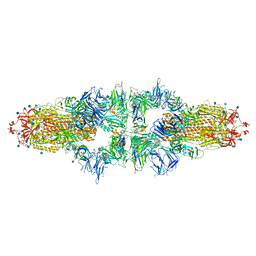 | | SARS-CoV-2 Spike (dimers) in complex with six Fu2 nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fu2 nanobody, Spike glycoprotein,Fibritin, ... | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo.
Nat Commun, 13, 2022
|
|
6KJ7
 
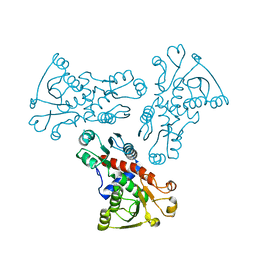 | | E. coli ATCase catalytic subunit mutant - G166P | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ9
 
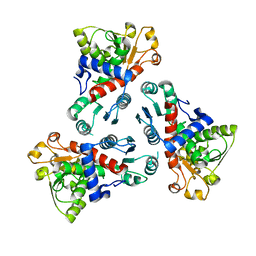 | | E. coli ATCase catalytic subunit mutant - G128/130A | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ8
 
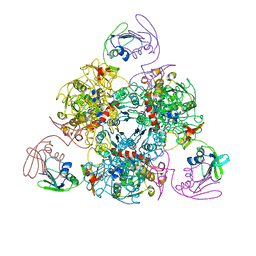 | | E. coli ATCase holoenzyme mutant - G166P (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
7M1C
 
 | |
7M30
 
 | |
7LYV
 
 | |
7M22
 
 | |
7LYW
 
 | |
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
8HLE
 
 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HJF
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 2 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJP
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJN
 
 | |
8HJO
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJG
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJQ
 
 | |
8HJL
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3E | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(3S,8S,9R,10S,11S,13R,14S,17S)-17-[(2S,5R)-5-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJH
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJK
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 1 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8IFO
 
 | | Crystal structure of estrogen related receptor-gamma DNA binding domain complexed with Pla2g12b promoter | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*CP*AP*AP*AP*GP*GP*TP*GP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*CP*AP*CP*CP*TP*TP*TP*GP*TP*CP*CP*TP*C)-3'), Estrogen-related receptor gamma, ... | | Authors: | Xu, T, Zhen, X, Liu, J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ERR gamma-DBD undergoes dimerization and conformational rearrangement upon binding to the downstream site of the DR1 element.
Biochem.Biophys.Res.Commun., 656, 2023
|
|
