5V3S
 
 | |
5VCR
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound uranium dioxide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, CHLORIDE ION, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VCS
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with Bound Acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
5WC7
 
 | | CypA Mutant - I97V S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
5XKU
 
 | | Crystal structure of hemagglutinin globular head from an H7N9 influenza virus in complex with a neutralizing antibody HNIgGA6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HNIgGA6 heavy chain, HNIgGA6 light chain, ... | | Authors: | Chen, C, Wang, J, Wang, W, Gao, X, Cui, S, Jin, Q. | | Deposit date: | 2017-05-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Insight into a Human Neutralizing Antibody against Influenza Virus H7N9
J. Virol., 92, 2018
|
|
3PE6
 
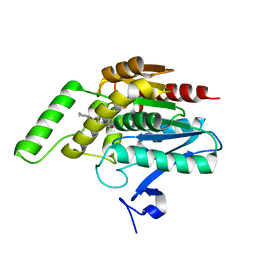 | |
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
4I80
 
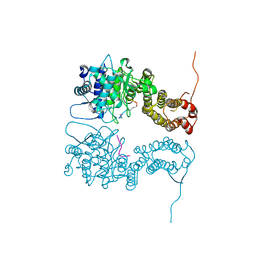 | |
2LN0
 
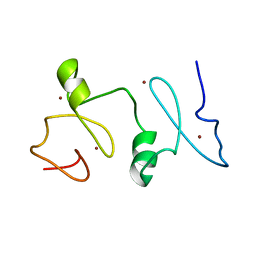 | | Structure of MOZ | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Qiu, Y. | | Deposit date: | 2011-12-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription.
Genes Dev., 26, 2012
|
|
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
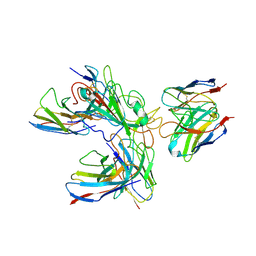 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
8UN5
 
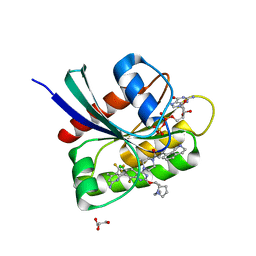 | | KRAS-G13D-GDP in complex with Cpd38 ((E)-1-((3S)-4-(7-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-6-chloro-8-fluoro-2-(((S)-2-methylenetetrahydro-1H-pyrrolizin-7a(5H)-yl)methoxy)quinazolin-4-yl)-3-methylpiperazin-1-yl)-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one) | | Descriptor: | (2E)-1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(4R,7aS)-2-methylidenetetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one, GLYCEROL, GTPase KRas, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8UN3
 
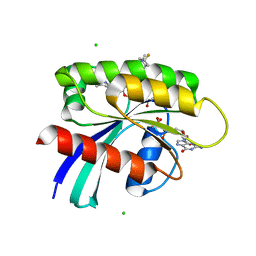 | | KRAS-G13D-GDP in complex with Cpd5 (1-((S)-10-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-11-chloro-7-(((2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl)methoxy)-3,4,13,13a-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-2(1H)-yl)prop-2-en-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(5M,8aS,13R)-5-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-2-{[(2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl]methoxy}-8a,9,11,12-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-10(8H)-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
7TQ4
 
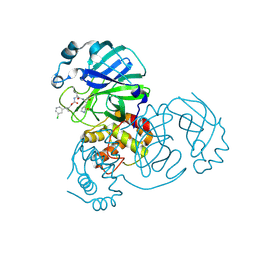 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
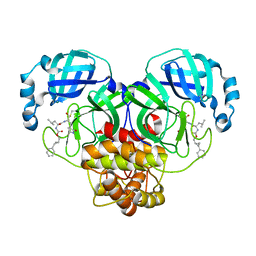 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
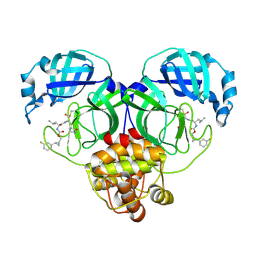 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8WE9
 
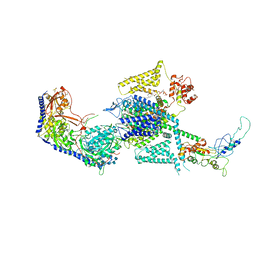 | | Human L-type voltage-gated calcium channel Cav1.2 (Class I) in the presence of pinaverium at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE8
 
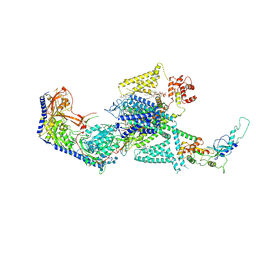 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine, amlodipine and pinaverium at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE7
 
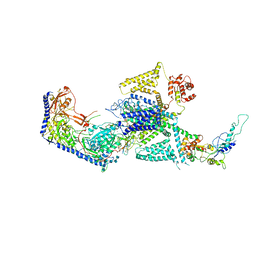 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
