3CE3
 
 | |
8TR3
 
 | | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CNE40 SOSIP Envelope glycoprotein gp120, ... | | Authors: | Chan, K.-W, Kong, X.P. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains.
Nat Commun, 15, 2024
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | Descriptor: | MERCURY (II) ION, Putative oxidoreductase | | Authors: | Yuan, Y.A, Yin, B. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
6CEZ
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V2 Fab 16C2 in complex with V2 peptide ConB | | Descriptor: | HIV-1 gp120 V2 Peptide Con B, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2, Light chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2 | | Authors: | Kong, X, Pan, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to alpha 4 beta 7.
PLoS Pathog., 14, 2018
|
|
3C1X
 
 | | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-MET in complex with a Pyrrolotriazine based inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-{[4-({5-[(4-aminopiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}oxy)-3-fluorophenyl]carbamoyl}-2-(4-fluorophenyl)acetamide | | Authors: | Sack, J. | | Deposit date: | 2008-01-24 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification of pyrrolo[2,1-f][1,2,4]triazine-based inhibitors of Met kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3AWD
 
 | | Crystal structure of gox2181 | | Descriptor: | CADMIUM ION, MAGNESIUM ION, Putative polyol dehydrogenase | | Authors: | Adam Yuan, Y, Yuan, Z. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analysis of Gox2181, a new member of the SDR superfamily from Gluconobacter oxydans.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
3CTH
 
 | |
6TQI
 
 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
2P4L
 
 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2007-03-12 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
3WUY
 
 | | Crystal structure of Nit6803 | | Descriptor: | Nitrilase | | Authors: | Yuan, Y.A, Yin, B, Wang, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
4GE5
 
 | |
4GE6
 
 | |
4GE2
 
 | |
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
7DOK
 
 | |
7DOI
 
 | |
5XXY
 
 | |
3F82
 
 | |
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DTO
 
 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
4BHK
 
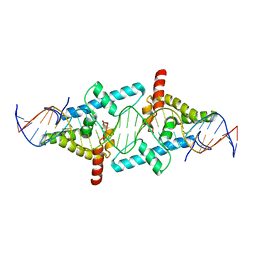 | | Crystal Structure of Moss Leafy bound to DNA | | Descriptor: | FLORICAULA/LEAFY HOMOLOG 1, MOSS-CR54 DNA | | Authors: | Nanao, M.H, Sayou, C, Dumas, R, Parcy, F. | | Deposit date: | 2013-04-03 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Promiscuous Intermediate Underlies the Evolution of Leafy DNA Binding Specificity.
Science, 343, 2014
|
|
8Q8J
 
 | | Crystal structure of human GPX4-R152H | | Descriptor: | Glutathione peroxidase | | Authors: | Napolitano, V, Mourao, A, Kolonko, M, Bostock, M.J, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human GPX4-R152H
To Be Published
|
|
8Q8N
 
 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
