8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
4U86
 
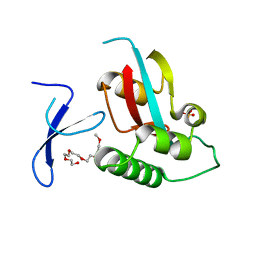 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U85
 
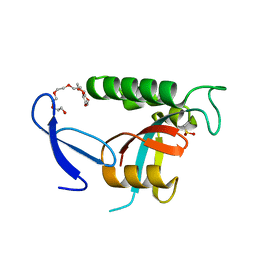 | | Human Pin1 with cysteine sulfinic acid 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U84
 
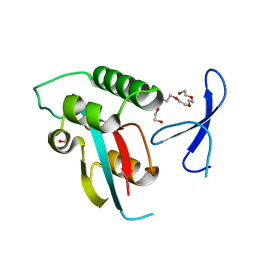 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
3NR9
 
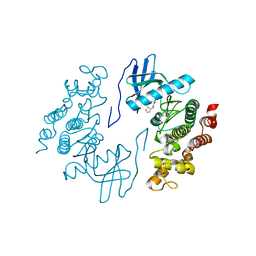 | | Structure of human CDC2-like kinase 2 (CLK2) | | Descriptor: | (5Z)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Dual specificity protein kinase CLK2 | | Authors: | Chaikuad, A, Savitsky, P, Krojer, T, Muniz, J.R.C, Filippakopoulos, P, Rellos, P, Keates, T, Fedorov, O, Pike, A.C.W, Eswaran, J, Berridge, G, Phillips, C, Zhang, Y, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of human CDC2-like kinase 2 (CLK2)
To be Published
|
|
3NPR
 
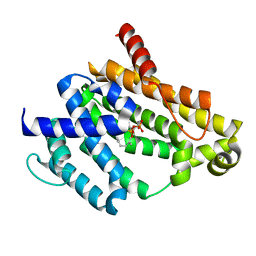 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from S. aureus complexed with Presqualene diphosphate (PSPP) | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Lin, F.-Y, Liu, C.-I, Liu, Y.-L, Wang, K, Zhang, Y, Oldfield, E. | | Deposit date: | 2010-06-28 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NRI
 
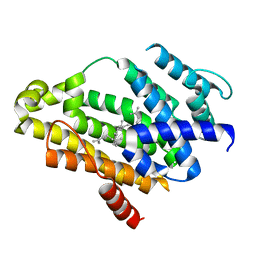 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from S. aureus complexed with dehydrosqualene (DHS) | | Descriptor: | (6E,10R,13S,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,11,13,14,18,22-octaene, Dehydrosqualene synthase | | Authors: | Lin, F.-Y, Liu, C.-I, Liu, Y.-L, Wang, K, Zhang, Y, Oldfield, E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2FF0
 
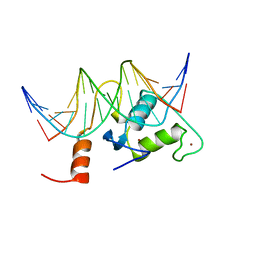 | | Solution Structure of Steroidogenic Factor 1 DNA Binding Domain Bound to its Target Sequence in the Inhibin alpha-subunit Promoter | | Descriptor: | CTGTGGCCCTGAGCC, GGCTCAGGGCCACAG, Steroidogenic factor 1, ... | | Authors: | Little, T.H, Zhang, Y, Matulis, C.K, Weck, J, Zhang, Z, Ramachandran, A, Mayo, K.E, Radhakrishnan, I. | | Deposit date: | 2005-12-17 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific deoxyribonucleic Acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability.
Mol.Endocrinol., 20, 2006
|
|
4EAC
 
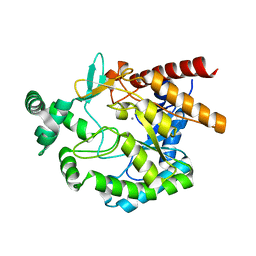 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
6LON
 
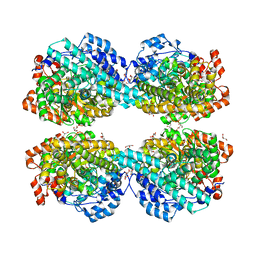 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8J86
 
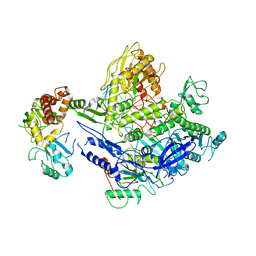 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*TP*)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-04-30 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8F
 
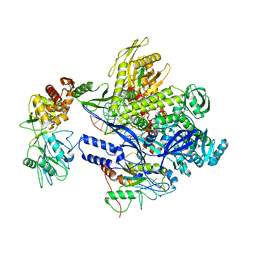 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8QQK
 
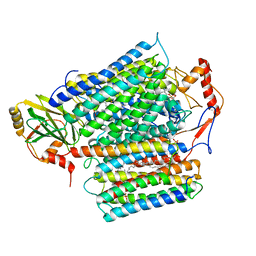 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8J8G
 
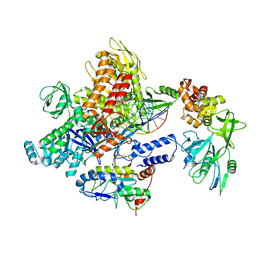 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
3OS5
 
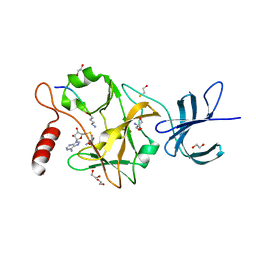 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1P9Z
 
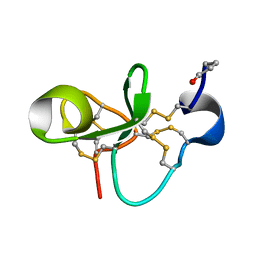 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
6V4X
 
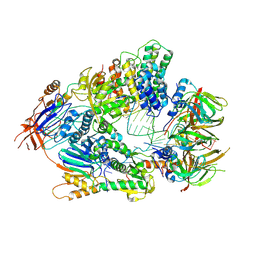 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
6VX7
 
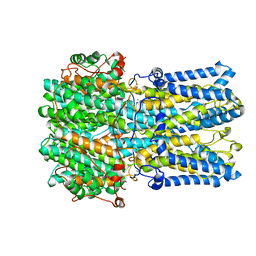 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5YPO
 
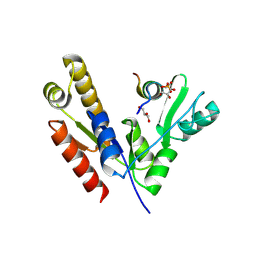 | | Crystal structure of PSD-95 GK domain in complex with phospho-SAPAP peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, SAPAP | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhang, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
6VX8
 
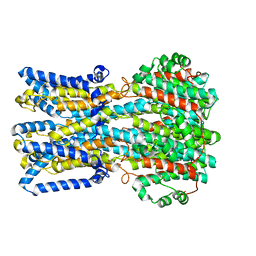 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
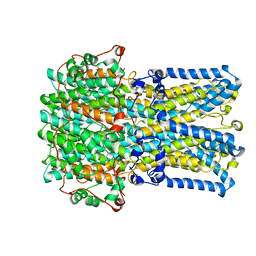 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5YPR
 
 | | Crystal Structure of PSD-95 SH3-GK domain in complex with a synthesized inhibitor | | Descriptor: | Disks large homolog 4, Synthesized GK inhibitor | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhu, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
