7EE3
 
 | | Crystal structure of PltC | | Descriptor: | Subtilase cytotoxin subunit B-like protein, TETRAETHYLENE GLYCOL | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE4
 
 | | Crystal structure of Neu5Ac bound PltC | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
3V3L
 
 | | Crystal structure of human RNF146 WWE domain in complex with iso-ADPRibose | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146 | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Genes Dev., 26, 2012
|
|
5KF4
 
 | |
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
4ILE
 
 | |
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
3EIQ
 
 | | Crystal structure of Pdcd4-eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | Authors: | Loh, P.G, Cheng, Z, Song, H. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
8YLC
 
 | | The crystal structure of PDE4D with Amentoflavone | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, 8-[5-[5,7-bis(oxidanyl)-4-oxidanylidene-chromen-2-yl]-2-oxidanyl-phenyl]-2-(4-hydroxyphenyl)-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.30003715 Å) | | Cite: | Discovery of amentoflavone as a natural PDE4 inhibitor with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
8GU5
 
 | | Wild type poly(ethylene terephthalate) hydrolase | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
8GU4
 
 | | Poly(ethylene terephthalate) hydrolase (IsPETase)-linker | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
5GMQ
 
 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
5H5Z
 
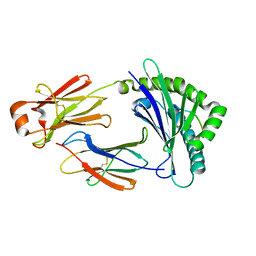 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
