2KKA
 
 | | Human telomere DNA two-tetrad quadruplex structure in K+ solution | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*IP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3' | | Authors: | Zhang, Z, Dai, J, Yang, D. | | Deposit date: | 2009-06-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures.
Nucleic Acids Res., 38, 2010
|
|
2MMU
 
 | | Structure of CrgA, a Cell Division Structural and Regulatory Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Das, N, Dai, J, Hung, I, Rajagopalan, M, Zhou, H, Cross, T.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MC7
 
 | | Structure of Salmonella MgtR | | Descriptor: | Regulatory peptide | | Authors: | Jean-Francois, F, Dai, J, Yu, L, Myrick, A, Rubin, E, Fajer, P, Song, L, Zhou, H, Cross, T. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study.
J.Mol.Biol., 426, 2014
|
|
5ZQZ
 
 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZRV
 
 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BSZ
 
 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3FM9
 
 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
3W2D
 
 | | Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment | | Descriptor: | Enterotoxin type B, Monoclonal Antibody 3E2 Fab figment heavy chain, Monoclonal Antibody 3E2 Fab figment light chain, ... | | Authors: | Liang, S.Y, Hu, S, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site.
MAbs, 6, 2014
|
|
1Y29
 
 | |
5T18
 
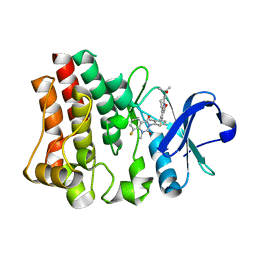 | | Crystal structure of Bruton agammabulinemia tyrosine kinase complexed with BMS-986142 aka (2s)-6-fluoro-5-[3-(8-fluoro-1-methyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-3-yl)-2-methylphenyl]-2-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1h-carbazole-8-carboxamide | | Descriptor: | 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-08-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton's Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers.
J. Med. Chem., 59, 2016
|
|
4DQO
 
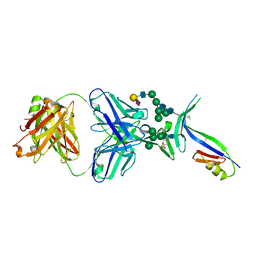 | | Crystal Structure of PG16 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | 1FD6-V1V2 scaffold ZM109 HIV-1 strain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 Fab Heavy Chain, ... | | Authors: | Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6O8I
 
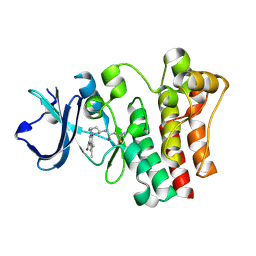 | | BTK In Complex With Inhibitor | | Descriptor: | 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Pokross, M, Tebben, A.J, Watterson, S.H. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of Branebrutinib (BMS-986195): A Strategy for Identifying a Highly Potent and Selective Covalent Inhibitor Providing Rapid in Vivo Inactivation of Bruton's Tyrosine Kinase (BTK).
J. Med. Chem., 62, 2019
|
|
8GPY
 
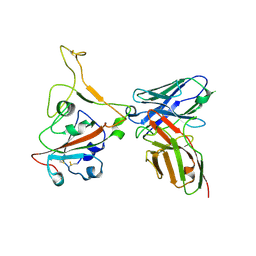 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
3CXD
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Yang, H, Zhong, C, Ding, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3F82
 
 | |
7KBV
 
 | |
7KBW
 
 | |
7KBX
 
 | |
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8JYS
 
 | |
5B7B
 
 | | Crystal structure of Nucleoprotein-nucleozin complex | | Descriptor: | Nucleoprotein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl](5-methyl-3-phenyl-1,2-oxazol-4-yl)methanone | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of H1N1 Nucleoprotein-Nucleozin Binding Sites
Sci Rep, 6, 2016
|
|
