7S8O
 
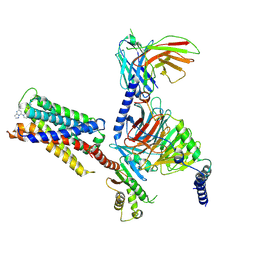 | | CryoEM structure of Gi-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8L
 
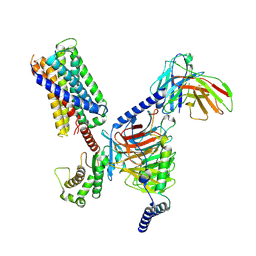 | | CryoEM structure of Gq-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8M
 
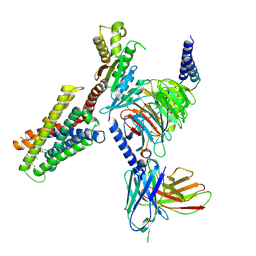 | | CryoEM structure of Gi-coupled MRGPRX2 with peptide agonist Cortistatin-14 | | Descriptor: | Cortistatin 14, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8N
 
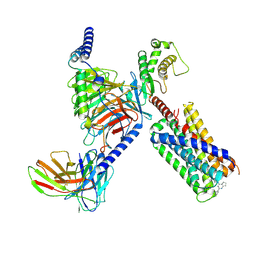 | | CryoEM structure of Gq-coupled MRGPRX2 with small molecule agonist (R)-Zinc-3573 | | Descriptor: | (3R)-N,N-dimethyl-1-[(8S)-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl]pyrrolidin-3-amine, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
7S8P
 
 | | CryoEM structure of Gq-coupled MRGPRX4 with small molecule agonist MS47134 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2021-09-18 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, function and pharmacology of human itch GPCRs.
Nature, 600, 2021
|
|
1P7M
 
 | | SOLUTION STRUCTURE AND BASE PERTURBATION STUDIES REVEAL A NOVEL MODE OF ALKYLATED BASE RECOGNITION BY 3-METHYLADENINE DNA GLYCOSYLASE I | | Descriptor: | 3-METHYL-3H-PURIN-6-YLAMINE, DNA-3-methyladenine glycosylase I, ZINC ION | | Authors: | Cao, C, Kwon, K, Jiang, Y.L, Drohat, A.C, Stivers, J.T. | | Deposit date: | 2003-05-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and base perturbation studies reveal a novel mode of alkylated base recognition by 3-methyladenine DNA glycosylase I
J.Biol.Chem., 278, 2003
|
|
5ZKP
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZKQ
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
9DQJ
 
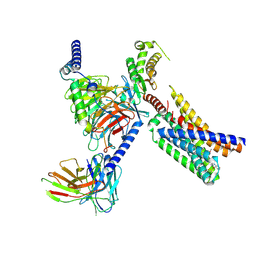 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-3945 | | Descriptor: | 2-({1-[1-(4-methoxyphenyl)cyclopropane-1-carbonyl]piperidin-4-yl}amino)quinazolin-4(3H)-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
9DQH
 
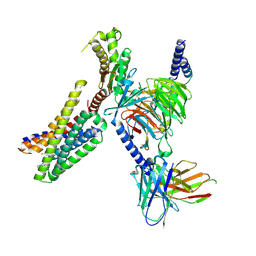 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-2825 | | Descriptor: | 2-({1-[2-(4-chlorophenyl)-2-methylpropanoyl]piperidin-4-yl}amino)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
1J0Q
 
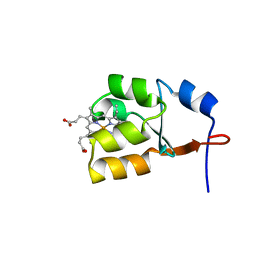 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
7SRS
 
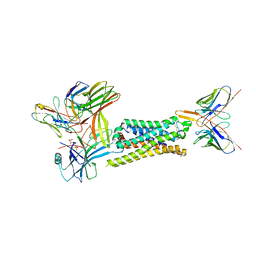 | | 5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B, ... | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7SRQ
 
 | | 5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7SRR
 
 | | 5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2B, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Cao, C, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
6K3K
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, DNA/RNA (5'-D(*AP*TP*TP*CP*UP*(ICY)P*AP*AP*TP*T)-3'), ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
6K3J
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product A | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*UP*(IUR)P*AP*AP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
7D3W
 
 | |
7D3X
 
 | |
8YRG
 
 | | CryoEM structure of fospropofol-bound MRGPRX4-Gq complex | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | MRGPRX4 mediates phospho-drug-associated pruritus in a humanized mouse model.
Sci Transl Med, 16, 2024
|
|
1WM8
 
 | | Solution Structure of BmP03 from the Venom of Scorpion Buthus martensii Karsch, 10 structures | | Descriptor: | Neurotoxin BmP03 | | Authors: | Wu, H, He, F, Li, Y, Wu, G, Cao, C, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of BmP03 from Venom of Scorpion Buthus martensii Karsch
Acta Chim.Sinica, 58, 2000
|
|
1NKU
 
 | |
2RIO
 
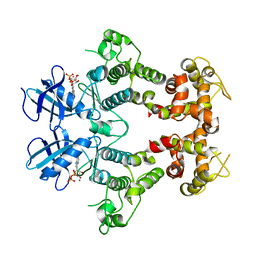 | | Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation of non-conventional splicing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, STRONTIUM ION, ... | | Authors: | Lee, K.P, Dey, M, Neculai, D, Cao, C, Dever, T.E, Sicheri, F. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the dual enzyme ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing.
Cell(Cambridge,Mass.), 132, 2008
|
|
1G7S
 
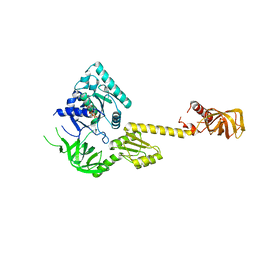 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G7R
 
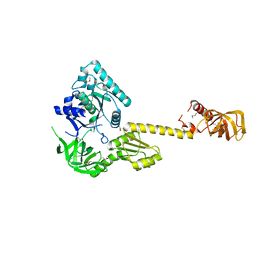 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B | | Descriptor: | TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1S0U
 
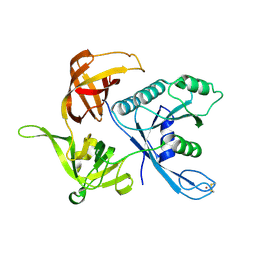 | | eIF2gamma apo | | Descriptor: | Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Roll-Mecak, A, Alone, P, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2004-01-04 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Structure of Translation Initiation Factor eIF2gamma: IMPLICATIONS FOR tRNA AND eIF2alpha BINDING.
J.Biol.Chem., 279, 2004
|
|
