8OZA
 
 | | Human cathepsin L in complex with covalently bound CA-074 methyl ester | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-05-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8OWL
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-6 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8PKP
 
 | | Cryo-EM structure of the apo Anaphase-promoting complex/cyclosome (APC/C) at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-07-19 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of apo-APC/C and APC/C CDH1:EMI1 complexes provide insights into APC/C regulation.
Nat Commun, 15, 2024
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8P8J
 
 | | Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8P3V
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P7W
 
 | | Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2 | | Descriptor: | 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ATP-binding cassette sub-family G member 2, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8PIV
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8OV0
 
 | | MUC5AC CysD7 amino acids 3518-3626 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mucin-5AC | | Authors: | Khmelnitsky, L, Milo, A, Dym, O, Fass, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity of CysD domains in gel-forming mucins.
Febs J., 290, 2023
|
|
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3U
 
 | |
8P3W
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3S
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P8A
 
 | | Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8PN6
 
 | | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus | | Descriptor: | Genome polyprotein, Serine protease subunit NS2B | | Authors: | Ni, X, Fairhead, M, Balcomb, B.H, Aschenbrenner, J.C, Ferreira, L.M, Godoy, A.S, Lithgo, R.M, MacLean, E.M, Marples, P.G, Thompson, W, Tomlinson, C.W.E, Szommer, T, Wild, C, Wright, N.D, Koekemoer, L, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus
To Be Published
|
|
8P3Q
 
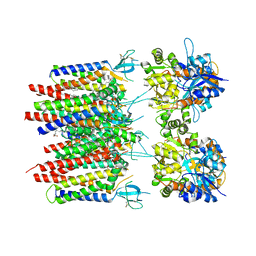 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Z
 
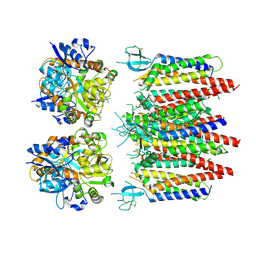 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Y
 
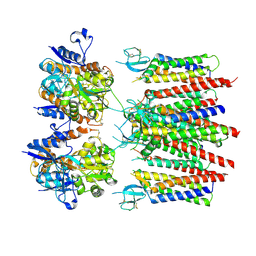 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
5HLP
 
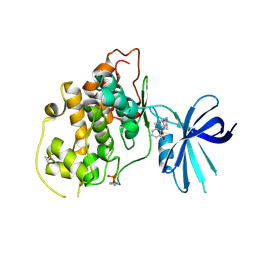 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
6W05
 
 | |
8PM6
 
 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
