2N1V
 
 | |
2LXP
 
 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2MDA
 
 | |
2MFK
 
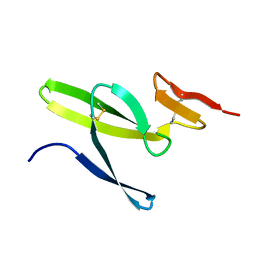 | |
7XGL
 
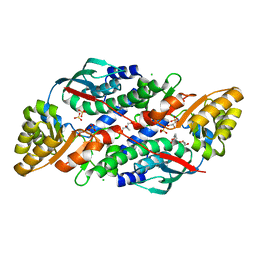 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in Apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGN
 
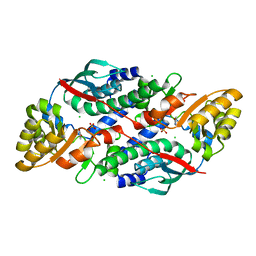 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Nicotinic Acid (NA) | | Descriptor: | CHLORIDE ION, NICOTINIC ACID, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
4E3C
 
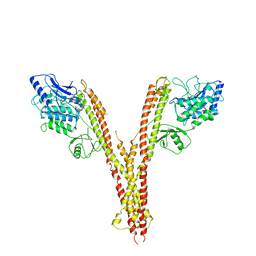 | | X-ray crystal structure of human IKK2 in an active conformation | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | Authors: | Polley, S, Huang, D.B, Hauenstein, A.V, Ghosh, G, Huxford, T. | | Deposit date: | 2012-03-09 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray crystal structure of human IKK2 in an active conformation
Plos.Biol., 2013
|
|
2JMH
 
 | | NMR solution structure of Blo t 5, a major mite allergen from Blomia tropicalis | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Naik, M.T, Chang, C, Kuo, I, Chua, K, Huang, T. | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Roles of Structure and Structural Dynamics in the Antibody Recognition of the Allergen Proteins: An NMR Study on Blomia tropicalis Major Allergen
Structure, 16, 2008
|
|
6QIP
 
 | |
6QIO
 
 | |
8YE6
 
 | |
8YE9
 
 | |
1KM8
 
 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
1KM9
 
 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
1M07
 
 | | RESIDUES INVOLVED IN THE CATALYSIS AND BASE SPECIFICITY OF CYTOTOXIC RIBONUCLEASE FROM BULLFROG (RANA CATESBEIANA) | | Descriptor: | 5'-D(*AP*CP*GP*A)-3', Ribonuclease | | Authors: | Leu, Y.-J, Chern, S.-S, Wang, S.-C, Hsiao, Y.-Y, Amiraslanov, I, Liaw, Y.-C, Liao, Y.-D. | | Deposit date: | 2002-06-12 | | Release date: | 2003-01-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and X-ray crystallography
J.Biol.Chem., 278, 2003
|
|
8EPA
 
 | |
8TGA
 
 | |
8TG9
 
 | |
