6KMH
 
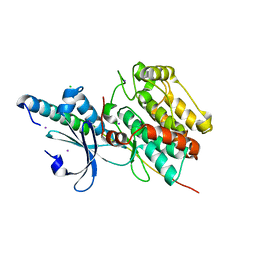 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
7D0Q
 
 | |
7D0P
 
 | |
7D0O
 
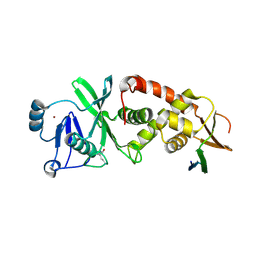 | | Crystal structure of human HBO1-BRPF2 in apo form | | Descriptor: | 1,2-ETHANEDIOL, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Li, W, Ding, J. | | Deposit date: | 2020-09-11 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | HBO1 is a versatile histone acyltransferase critical for promoter histone acylations.
Nucleic Acids Res., 49, 2021
|
|
7D0R
 
 | |
7D0S
 
 | |
4MPL
 
 | | Crystal structure of BMP9 at 1.90 Angstrom | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Li, W, Morrell, N.W, Wei, Z. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of Bone Morphogenetic Protein 9 (BMP9) by Redox-dependent Proteolysis.
J.Biol.Chem., 289, 2014
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XSJ
 
 | | The structure of the Mint1/Munc18-1/syntaxin-1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Feng, W, Li, W. | | Deposit date: | 2022-05-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A non-canonical target-binding site in Munc18-1 domain 3b for assembling the Mint1-Munc18-1-syntaxin-1 complex.
Structure, 31, 2023
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7E4Z
 
 | | Crystal structure of tubulin in complex with Maytansinol | | Descriptor: | (1R,2S,3S,5S,6S,16E,18E,20R,21S)-11-chloro-6,21-dihydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4Q
 
 | | Crystal structure of tubulin in complex with L-DM1-SMe | | Descriptor: | (1S,2R,3R,5R,6R,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methyldisulfanyl)propanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4R
 
 | | Crystal structure of tubulin in complex with D-DM1-SMe | | Descriptor: | (10E,12E)-86-chloro-14-hydroxy-85,14-dimethoxy-33,2,7,10-tetramethyl-12,6-dioxo-7-aza-1(6,4)-oxazinana-3(2,3)-oxirana-8(1,3)-benzenacyclotetradecaphane-10,12-dien-4-yl N-methyl-N-(3-(methylsulfinothioyl)propanoyl)-D-alaninate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7WL4
 
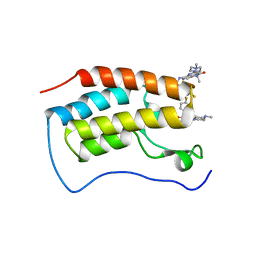 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor SLP-50 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-ethyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]-2,4-bis(fluoranyl)benzenesulfonamide | | Authors: | Zhang, C, Wang, C, Li, W, Zhang, Y, Xu, Y, Sun, L. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis, and anticancer evaluation of ammosamide B with pyrroloquinoline derivatives as novel BRD4 inhibitors.
Bioorg.Chem., 127, 2022
|
|
7W30
 
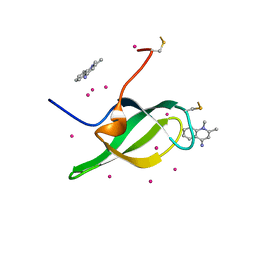 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7W2P
 
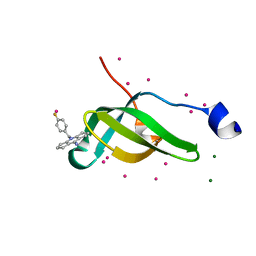 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 2-[(4-fluorophenyl)methyl]-2-azatricyclo[7.3.0.0^{3,7}]dodeca-1(9),3(7)-dien-8-imine, MAGNESIUM ION, Survival motor neuron protein, ... | | Authors: | Li, W, Arrowsmith, C.H, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
5OWD
 
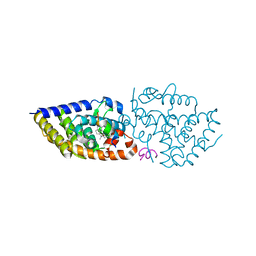 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
5OW7
 
 | | VDR complex | | Descriptor: | (3~{S})-3-[(1~{S},3~{a}~{S},4~{E},7~{a}~{S})-7~{a}-methyl-4-[(2~{Z})-2-[(5~{S})-2-methylidene-5-oxidanyl-cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-3-oxidanyl-~{N}-propan-2-yl-butanamide, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
5OW9
 
 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-heptan-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
6WVA
 
 | |
1MR1
 
 | | Crystal Structure of a Smad4-Ski Complex | | Descriptor: | Mothers against decapentaplegic homolog 4, Ski oncogene, ZINC ION | | Authors: | Wu, J.-W, Krawitz, A.R, Chai, J, Li, W, Zhang, F, Luo, K, Shi, Y. | | Deposit date: | 2002-09-17 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-mediated Repression of TGF-beta Signaling
Cell(Cambridge,Mass.), 111, 2002
|
|
6XE0
 
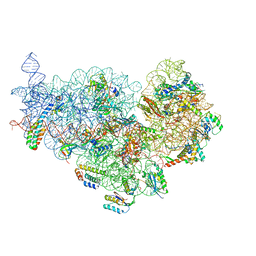 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
2GD4
 
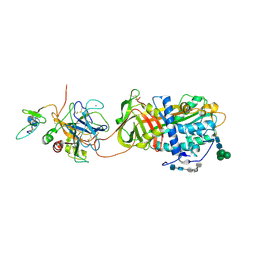 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
5XI7
 
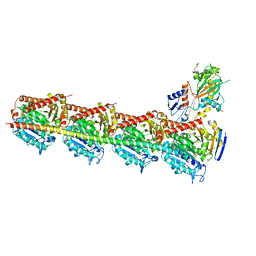 | | Crystal structure of T2R-TTL bound with PO-7 | | Descriptor: | (6Z)-3-[[2,5-bis(fluoranyl)phenyl]methylidene]-6-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Chu, Y, Wang, Y, Yang, J, Li, W. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
