4DS7
 
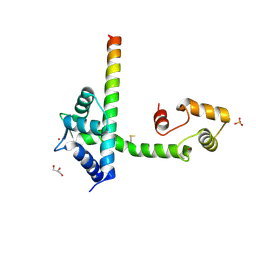 | |
7XUE
 
 | |
4UWY
 
 | | FGFR1 Apo structure | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Thiyagarajan, N, Bunney, T, Katan, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3P2X
 
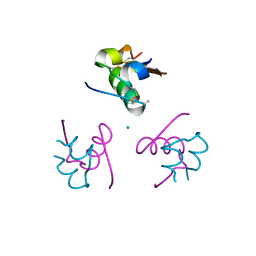 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3P33
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
6AX1
 
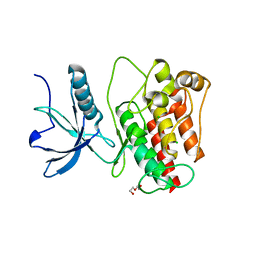
 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
3BXQ
 
 | | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition | | Descriptor: | ZINC ION, insulin A chain, insulin B chain | | Authors: | Wan, Z.L, Huang, K, Hu, S.Q, Whittaker, J, Weiss, M.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition.
J.Biol.Chem., 283, 2008
|
|
3L13
 
 | | Crystal Structures of Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors | | Descriptor: | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, [3-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenyl]methanol | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3L17
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 4-methyl-5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyrimidin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
1XW7
 
 | | Diabetes-Associated Mutations in Human Insulin: Crystal Structure and Photo-Cross-Linking Studies of A-Chain Variant Insulin Wakayama | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Huang, K, Xu, B, Chu, Y.C, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-10-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Diabetes-associated mutations in human insulin: crystal structure and photo-cross-linking studies of a-chain variant insulin wakayama
Biochemistry, 44, 2005
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3V19
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3V1G
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3FQ9
 
 | | Design of an insulin analog with enhanced receptor-binding selectivity. Rationale, structure, and therapeutic implications | | Descriptor: | Insulin, ZINC ION | | Authors: | Zhao, M, Wan, Z.L, Whittaker, L, Xu, B, Phillips, N, Katsoyannis, P, Whittaker, J, Weiss, M.A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of an insulin analog with enhanced receptor binding selectivity: rationale, structure, and therapeutic implications.
J.Biol.Chem., 284, 2009
|
|
3GKY
 
 | | The Structural Basis of an ER Stress-Associated Bottleneck in a Protein Folding Landscape | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Liu, M, Wan, Z.L, Chu, Y.C, Alddin, H, Klaproth, B, Weiss, M.A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a "nonfoldable" insulin: IMPAIRED FOLDING EFFICIENCY DESPITE NATIVE ACTIVITY.
J.Biol.Chem., 284, 2009
|
|
4F2L
 
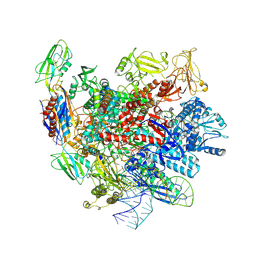
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
3T8M
 
 | |
2M5H
 
 | |
6K2U
 
 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
3TL5
 
 | | Discovery of GDC-0980: a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor for the Treatment of Cancer | | Descriptor: | (2S)-1-(4-{[2-(2-aminopyrimidin-5-yl)-7-methyl-4-(morpholin-4-yl)thieno[3,2-d]pyrimidin-6-yl]methyl}piperazin-1-yl)-2-hydroxypropan-1-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-08-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor (GDC-0980) for the Treatment of Cancer.
J.Med.Chem., 54, 2011
|
|
3NZU
 
 | |
3NZS
 
 | |
4W2R
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
