3K53
 
 | | Crystal Structure of NFeoB from P. furiosus | | Descriptor: | Ferrous iron transport protein b | | Authors: | Eng, E.T, Dong, G, Unger, V.M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural fold, conservation and Fe(II) binding of the intracellular domain of prokaryote FeoB.
J.Struct.Biol., 170, 2010
|
|
8FLJ
 
 | | Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA | | Descriptor: | CRISPR leader and repeat, anti-sense strand of DNA, CRISPR leader, ... | | Authors: | Santiago-Frangos, A, Henriques, W.S, Wiegand, T, Gauvin, C, Buyukyoruk, M, Neselu, K, Eng, E.T, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PXM
 
 | | Horse spleen apoferritin light chain | | Descriptor: | Ferritin light chain | | Authors: | Kopylov, M, Kelley, K, Yen, L.Y, Rice, W.J, Eng, E.T, Carragher, B, Potter, C.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Horse spleen apoferritin light chain structure at 2.1 Angstrom resolution
To Be Published
|
|
7S0F
 
 | | Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi protein | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Paknejad, N, Alegre, K.O, Su, M, Lou, J.S, Huang, J, Jordan, K.D, Eng, E.T, Meyerson, J.R, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and mechanism of activation of two different families of G proteins by the same GPCR.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7S0G
 
 | | Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi/s chimera protein | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Paknejad, N, Alegre, K.O, Su, M, Lou, J.S, Huang, J, Jordan, K.D, Eng, E.T, Meyerson, J.R, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis and mechanism of activation of two different families of G proteins by the same GPCR.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6WIV
 
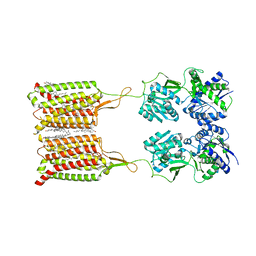 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
6MUR
 
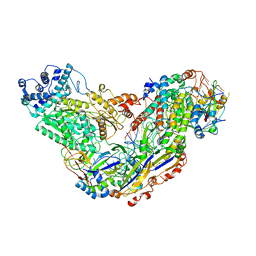 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MUT
 
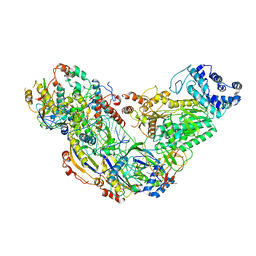 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MUU
 
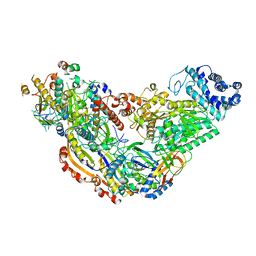 | | Cryo-EM structure of Csm-crRNA binary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (25-MER), Uncharacterized protein Csm1, Uncharacterized protein Csm2, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MUS
 
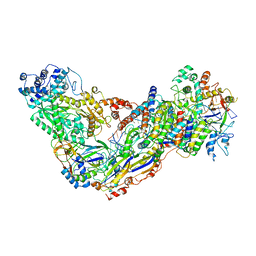 | | Cryo-EM structure of larger Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (25-MER), RNA (33-MER), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6BBM
 
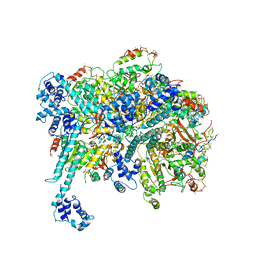 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | Authors: | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | Deposit date: | 2017-10-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
7RDX
 
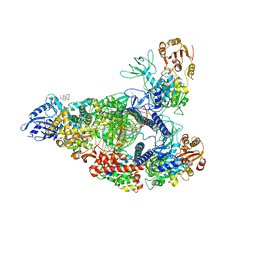 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
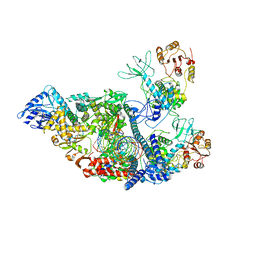 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
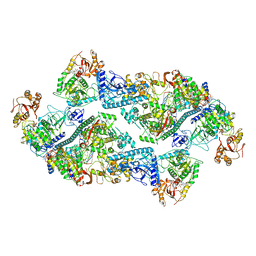 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
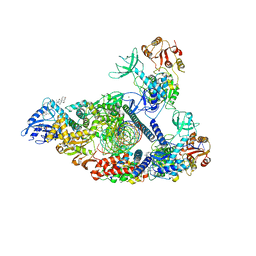 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
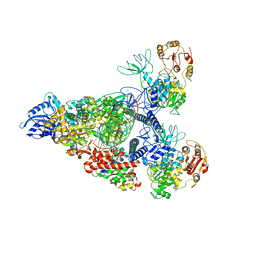 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
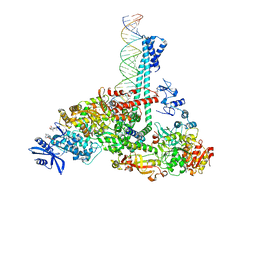 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6OT1
 
 | |
6OSY
 
 | |
6XEZ
 
 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
