4HC4
 
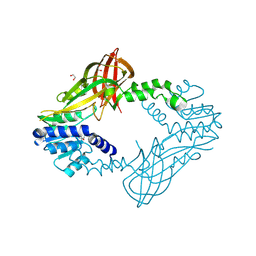 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4H12
 
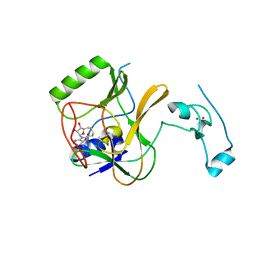 | | The crystal structure of methyltransferase domain of human SET domain-containing protein 2 in complex with S-adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Amaya, M.F, Dong, A, Zeng, H, Mackenzie, F, Bunnage, M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
8R6A
 
 | |
8R7O
 
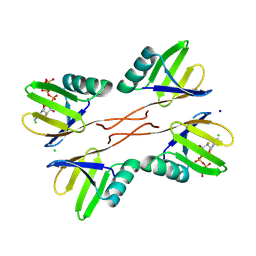 | | HUWE1 WWE domain in complex with 2'F-ATP | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1, SODIUM ION, ... | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8RD1
 
 | | HUWE1 WWE domain in complex with compound 4 | | Descriptor: | 2-(2-hydroxy-2-oxoethyl)-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8RD7
 
 | | HUWE1 WWE domain in complex with ADP-ribose | | Descriptor: | ACETATE ION, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1, ... | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8R5N
 
 | | DTX1 WWE domain in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase DTX1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8R6B
 
 | |
8RD0
 
 | | HUWE1 WWE domain in complex with compound 3 | | Descriptor: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6TSZ
 
 | | The ULK4 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ULK4 | | Authors: | Preuss, F, Chatterjee, D, Mathea, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide Binding, Evolutionary Insights, and Interaction Partners of the Pseudokinase Unc-51-like Kinase 4.
Structure, 28, 2020
|
|
6TSG
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
6TU9
 
 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
6TUA
 
 | | The RYK Pseudokinase Domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase RYK | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
6TXS
 
 | | The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide | | Descriptor: | CD44 antigen, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6TXQ
 
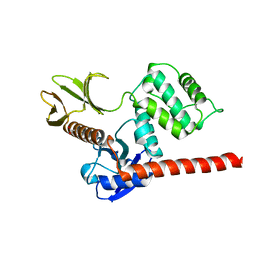 | | The high resolution structure of the FERM domain and helical linker of human moesin | | Descriptor: | ACETATE ION, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6V2H
 
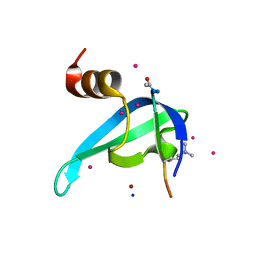 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2S
 
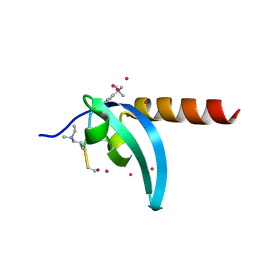 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6VDB
 
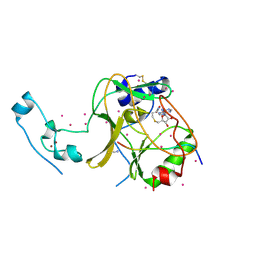 | | SETD2 in complex with a H3-variant super-substrate peptide | | Descriptor: | ALA-PRO-ARG-PHE-GLY-GLY-VAL-MET-ARG-PRO-ASN-ARG, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Beldar, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Jeltsch, A, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence specificity analysis of the SETD2 protein lysine methyltransferase and discovery of a SETD2 super-substrate.
Commun Biol, 3, 2020
|
|
6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V8W
 
 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V9T
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
