2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2MZY
 
 | | 1H, 13C, and 15N Chemical Shift Assignments and structure of Probable Fe(2+)-trafficking protein from Burkholderia pseudomallei 1710b. | | Descriptor: | Probable Fe(2+)-trafficking protein | | Authors: | Tang, C, Yang, F, Barnwal, R, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Probable Fe(2+)-trafficking protein
To be Published
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N7B
 
 | | Solution structure of the human Siglec-8 lectin domain in complex with 6'sulfo sialyl Lewisx | | Descriptor: | 3-aminopropan-1-ol, N-acetyl-alpha-neuraminic acid-(2-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1VIW
 
 | | TENEBRIO MOLITOR ALPHA-AMYLASE-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, ALPHA-AMYLASE-INHIBITOR, ... | | Authors: | Nahoum, V, Egloff, M.P, Payan, F. | | Deposit date: | 1998-07-21 | | Release date: | 1999-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A plant-seed inhibitor of two classes of alpha-amylases: X-ray analysis of Tenebrio molitor larvae alpha-amylase in complex with the bean Phaseolus vulgaris inhibitor.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2Y
 
 | |
7WZ9
 
 | | HSA-In agent complex | | Descriptor: | 16-chloranyl-~{N},~{N}-dimethyl-15-thia-1$l^{4},12$l^{4},13-triaza-16$l^{4}-indatetracyclo[8.6.0.0^{2,7}.0^{12,16}]hexadeca-1,3,5,7,9,11,13-heptaen-14-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, Z.L, Yang, F. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of HSA-In agent complex
To Be Published
|
|
7Y2D
 
 | | HSA-Cu agent complex | | Descriptor: | 13-methoxy-~{N},~{N}-dimethyl-2-oxa-4-thia-6,7$l^{4}-diaza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(9),5,7,10,12-pentaen-5-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, Z.L, Yang, F. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HSA-Cu agent complex
To Be Published
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
7CFS
 
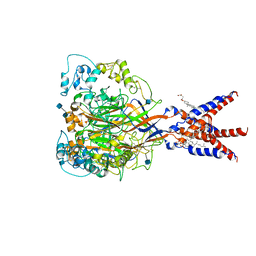 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7XAV
 
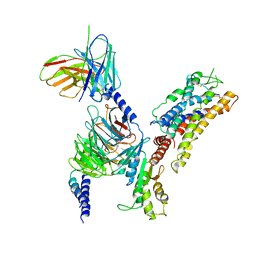 | | Structure of somatostatin receptor 2 bound with lanreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAT
 
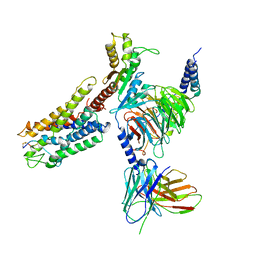 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
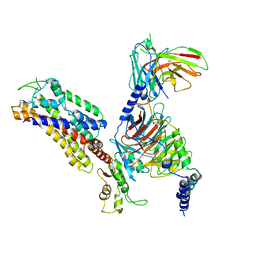 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7CFT
 
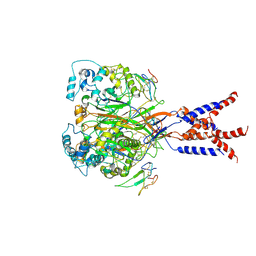 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7D72
 
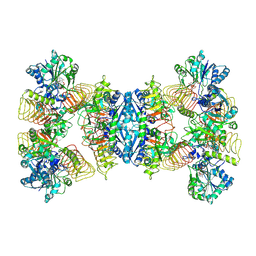 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
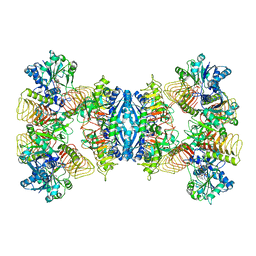 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
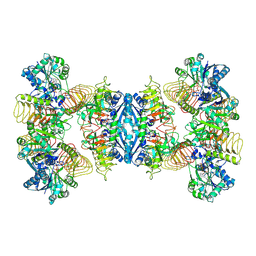 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7XWO
 
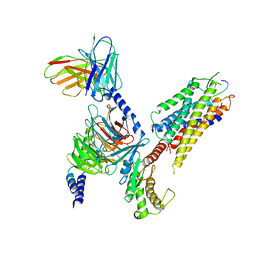 | | Neurokinin A bound to active human neurokinin 2 receptor in complex with G324 | | Descriptor: | G324, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W.J, Yuan, Q.N, Zhang, H.H, Yang, F, Ling, S.L, Lv, P, Eric, X, Tian, C.L, Yin, W.C, Shi, P. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the activation of neurokinin 2 receptor by neurokinin A.
Cell Discov, 8, 2022
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2CY3
 
 | |
6K2U
 
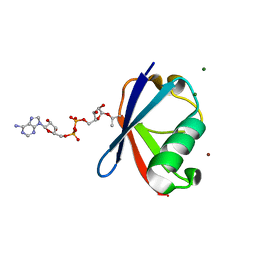 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
7X0L
 
 | |
7X0H
 
 | |
7X0P
 
 | |
