8XMM
 
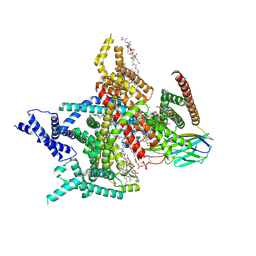 | | Voltage-gated sodium channel Nav1.7 variant M9 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YB6
 
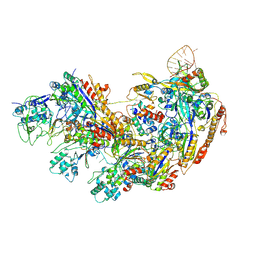 | | Type I-EHNH Cascade complex | | Descriptor: | 61-nt crRNA, CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YEO
 
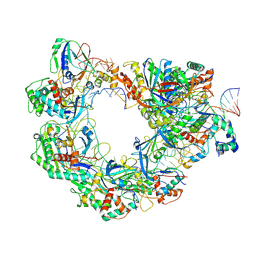 | | Type I-FHNH Cascade-dsDNA R-loop complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YDB
 
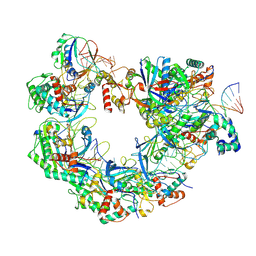 | | Type I-FHNH Cascade-dsDNA intermediate complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YH9
 
 | | Type I-FHNH Cascade complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YHA
 
 | | Type I-EHNH Cascade-ssDNA complex | | Descriptor: | 61-nt crRNA, CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
7DTC
 
 | | voltage-gated sodium channel Nav1.5-E1784K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Pan, X, Li, Z. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-24 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na v 1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
4LAK
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Wang, R, Li, Z, Xu, G.L, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
1TVI
 
 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | Descriptor: | Hypothetical UPF0054 protein TM1509 | | Authors: | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
3J6J
 
 | | 3.6 Angstrom resolution MAVS filament generated from helical reconstruction | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Wu, B, Peisley, A, Li, Z, Egelman, E, Walz, T, Penczek, P, Hur, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I.
Mol.Cell, 55, 2014
|
|
3SHB
 
 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
9BP9
 
 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, dimer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
9BPA
 
 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, tetramer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
5XWP
 
 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
5C6H
 
 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
3ZIU
 
 | | Crystal structure of Mycoplasma mobile Leucyl-tRNA Synthetase with Leu-AMS in the active site | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, LEUCYL-TRNA SYNTHETASE | | Authors: | Li, L, Palencia, A, Lukk, T, Li, Z, Luthey-Schulten, Z.A, Cusack, S, Martinis, S.A, Boniecki, M.T. | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leucyl-tRNA Synthetase Editing Domain Functions as a Molecular Rheostat to Control Codon Ambiguity in Mycoplasma Pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6TYY
 
 | | Hedgehog autoprocessing mutant D46H | | Descriptor: | Protein hedgehog | | Authors: | Li, H, Li, Z, Wang, C, Callahan, B.P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing.
J.Am.Chem.Soc., 141, 2019
|
|
