7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
8JCB
 
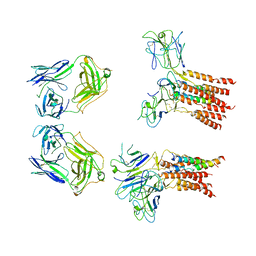 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JC0
 
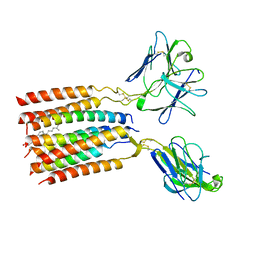 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JT3
 
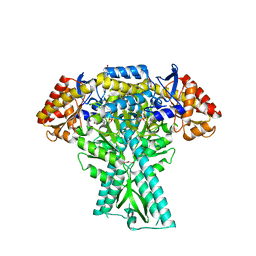 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, ACETATE ION, CrmG, ... | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2023-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Co-crystal structure provides insights on transaminase CrmG recognition amino donor L-Arg.
Biochem.Biophys.Res.Commun., 675, 2023
|
|
7DLY
 
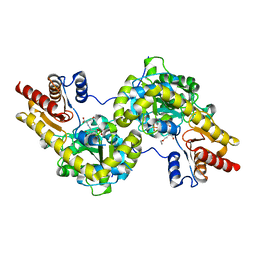 | | Crystal structure of Arabidopsis ACS7 mutant in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7 | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DLW
 
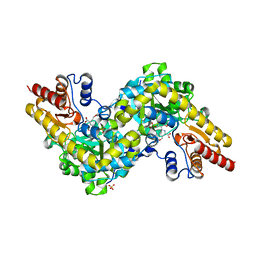 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7VLB
 
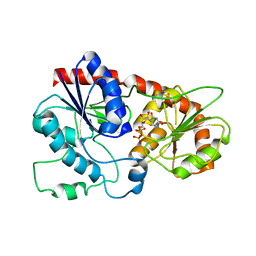 | |
8H7Q
 
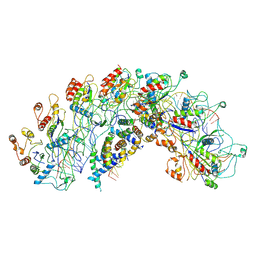 | | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution
To Be Published
|
|
8H67
 
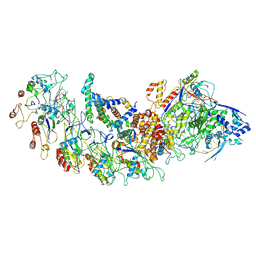 | | type I-B Cascade bound to a PAM-containing dsDNA target at 3.8 angstrom resolution. | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome editing of type I-B CRISPR-Cas.
Nat Commun, 15, 2024
|
|
7WDL
 
 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
7WL4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor SLP-50 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-ethyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]-2,4-bis(fluoranyl)benzenesulfonamide | | Authors: | Zhang, C, Wang, C, Li, W, Zhang, Y, Xu, Y, Sun, L. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis, and anticancer evaluation of ammosamide B with pyrroloquinoline derivatives as novel BRD4 inhibitors.
Bioorg.Chem., 127, 2022
|
|
7XZO
 
 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
 | |
7XZP
 
 | |
7YL2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
7E7E
 
 | | The co-crystal structure of ACE2 with Fab | | Descriptor: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7E3J
 
 | |
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8GZG
 
 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc
Proc.Natl.Acad.Sci.USA, 2023
|
|
8GZH
 
 | |
3T0K
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-20 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
