4Z8B
 
 | | crystal structure of a DGL mutant - H51G H131N | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, GLYCEROL, ... | | Authors: | Zamora-Caballero, S, Perez, A, Sanz, L, Bravo, J, Calvete, J.J. | | Deposit date: | 2015-04-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Quaternary structure of Dioclea grandiflora lectin assessed by equilibrium sedimentation and crystallographic analysis of recombinant mutants.
Febs Lett., 589, 2015
|
|
4Z92
 
 | | crystal structure of parechovirus-1 virion | | Descriptor: | Capsid subunit VP3, RNA (5'-R(*AP*UP*UP*UP*UP*U)-3'), capsid subunit VP0, ... | | Authors: | Kalynych, S, Palkova, L, Plevka, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid.
J.Virol., 90, 2015
|
|
5GJK
 
 | | Crystal Structure of BAF47 and BAF155 Complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, SWI/SNF complex subunit SMARCC1, ... | | Authors: | Yan, L, Qian, C. | | Deposit date: | 2016-06-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural Insights into BAF47 and BAF155 Complex Formation.
J. Mol. Biol., 429, 2017
|
|
7NBI
 
 | |
7NL5
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1R,2R,3R,4S,5R)-4-(hydroxymethyl)cyclohexane-1,2,3,5-tetrol, (1R,6S)-5beta-(Hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2beta,3beta,4alpha-triol, Alpha-1,6-mannanase, ... | | Authors: | Schroeder, S, Offen, W.A, Males, A, Jin, Y, De Boer, C, Enotarpi, J, Marino, L, van der Marel, G.A, Florea, B.I, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
4ZKR
 
 | |
5GVT
 
 | |
7NJJ
 
 | | Proteinase K grown inside HARE serial crystallography chip | | Descriptor: | NITRATE ION, Proteinase K | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NKF
 
 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4ZKS
 
 | |
7NJG
 
 | | Xylose isomerase grown inside HARE serial crystallography chip | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJI
 
 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5GT7
 
 | | Crystal Structure of Arg-bound CASTOR1 | | Descriptor: | ARGININE, GATS-like protein 3, MALONATE ION | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
5H3E
 
 | | Crystal structure of mouse isocitrate dehydrogenases 2 K256Q mutant complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
4ZQN
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | Descriptor: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
7N8C
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-06-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7N89
 
 | |
4Z0V
 
 | | The structure of human PDE12 residues 161-609 | | Descriptor: | 2',5'-phosphodiesterase 12, GLYCEROL, MAGNESIUM ION | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
5H1C
 
 | | Human RAD51 post-synaptic complexes | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
3Q1Y
 
 | |
5H3F
 
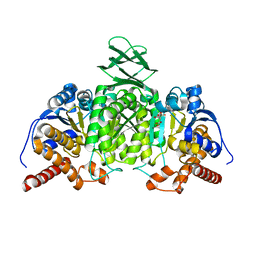 | | Crystal structure of mouse isocitrate dehydrogenases 2 complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
5H2T
 
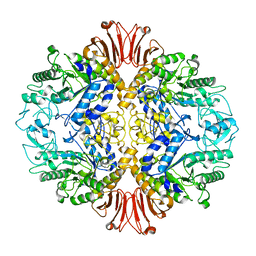 | |
5H4Y
 
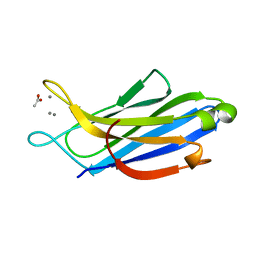 | | Crystal structure of human synaptotagmin 5 C2A domain | | Descriptor: | ACETATE ION, CALCIUM ION, Synaptotagmin-5 | | Authors: | Qiu, X, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2016-11-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of Ca(2+)-binding pocket of synaptotagmin 5 C2A domain
Int. J. Biol. Macromol., 95, 2017
|
|
5H4P
 
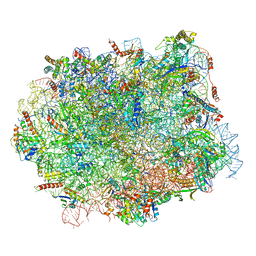 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
