6DAT
 
 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
6E5G
 
 | |
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
8EGZ
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EH0
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate and complexed with quinoline N-oxide | | Descriptor: | 1,2-ETHANEDIOL, 1-oxo-1lambda~5~-quinoline, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EH1
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate and complexed with 4-hydroxyquinoline | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Engineered tyrosine synthase (TmTyrS1), POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EUO
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
8EGY
 
 | | Engineered holo tyrosine synthase (TmTyrS1) derived from T. maritima TrpB | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
8ERX
 
 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
8EGK
 
 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
6TYF
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYG
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
6U0O
 
 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PPM
 
 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPN
 
 | | SHP2 catalytic domain in complex with CD28 (183-198) phosphopeptide (pTyr-191, p-Thr-195) | | Descriptor: | GLYCEROL, T-cell-specific surface glycoprotein CD28, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPL
 
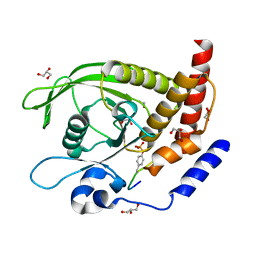 | | SHP2 catalytic domain in complex with IRS1 (625-639) phosphopeptide (pTyr-632, pSer-636) | | Descriptor: | ETHANOL, GLYCEROL, Insulin receptor substrate 1, ... | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
6TYE
 
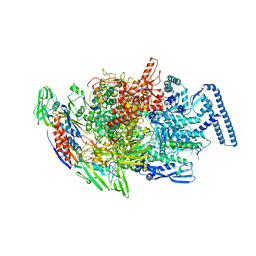 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PUX
 
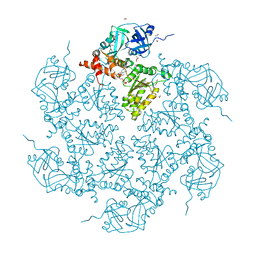 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
7PV9
 
 | |
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
